Inferring chemical reaction patterns using rule composition in graph grammars
Jakob L Andersen, Christoph Flamm, Daniel Merkle, Peter F Stadler
J Sys Chem 4:4 (2013), doi:10.1186/1759-2208-4-4 |
A graph-based toy model of chemistry
Gil Benkö, Christoph Flamm, Peter F Stadler
In Journal of Chemical Information and Computer Sciences, 2003, 43, 4, 1085-1093
doi:10.1021/ci0200570 bibtex |
Evolution of metabolic networks: a computational framework
Christoph Flamm, Alexander Ullrich, Heinz Ekker, Martin Mann, Daniel Högerl, Markus Rohrschneider, Sebastian Sauer, Gerik Scheuermann, Konstantin Klemm, Ivo L Hofacker, Peter F Stadler
In Journal of Systems Chemistry, 2010, 1, 1, 1-14
doi:10.1186/1759-2208-1-4 bibtex |
The SBML ODE Solver Library: a native API for symbolic and fast numerical analysis of reaction networks
Rainer Machné, Andrew Finney, Stefan Müller, James Lu, Stefanie Widder, Christoph Flamm
In Bioinformatics, 2006, 22, 11, 1406-1407
doi:10.1093/bioinformatics/btl086 bibtex |

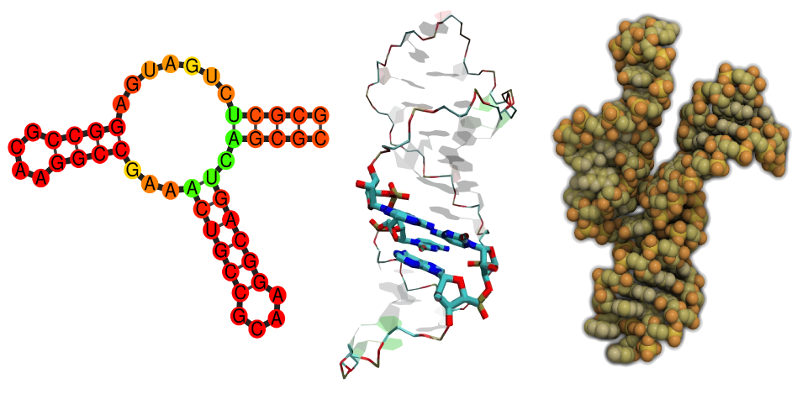 The development of algorithms to detect RNA genes, the
prediction of their structure on different levels of
abstraction, as well as the detailed characterization of
these molecules form an essential topic in our research
group.
The development of algorithms to detect RNA genes, the
prediction of their structure on different levels of
abstraction, as well as the detailed characterization of
these molecules form an essential topic in our research
group. 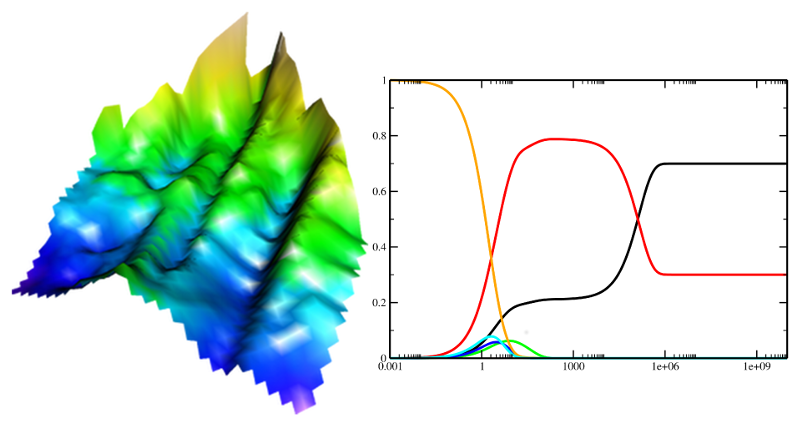 Strongly connected to RNA computational biology studies
is the investigation of RNA folding behaviors and
dynamics. The application and development of methods to
study such processes in terms of the underlying energy
landscapes is a challenging but successful part of our
research.
Strongly connected to RNA computational biology studies
is the investigation of RNA folding behaviors and
dynamics. The application and development of methods to
study such processes in terms of the underlying energy
landscapes is a challenging but successful part of our
research.
 The lessons learned from computational RNA biology and
folding dynamics allow us to develop methods to specifically design small
regulatory RNAs with a variety of functions, such as
ribozymes or riboswitches.
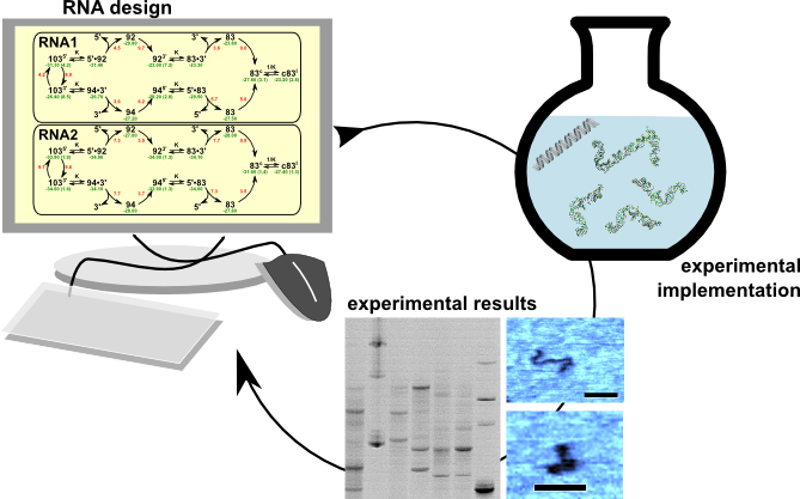
The lessons learned from computational RNA biology and
folding dynamics allow us to develop methods to specifically design small
regulatory RNAs with a variety of functions, such as
ribozymes or riboswitches.
 Modeling chemical reaction networks allows us to analyze all possible
reactions within a system and their interconnection via
different substrates.
We developed an efficient graph grammar approach to
describe these changes in connectivity within such networks
over time while cascades of chemical reactions take
place. Using such a formalism permits abstract reasoning about
chemical reaction networks and allows us to identify reaction
mechanisms underlying complex phenomena. We apply our methodology to a
broad set of (bio)chemical problems such as synthesis
planning or the evolution of catalyzed reaction networks.
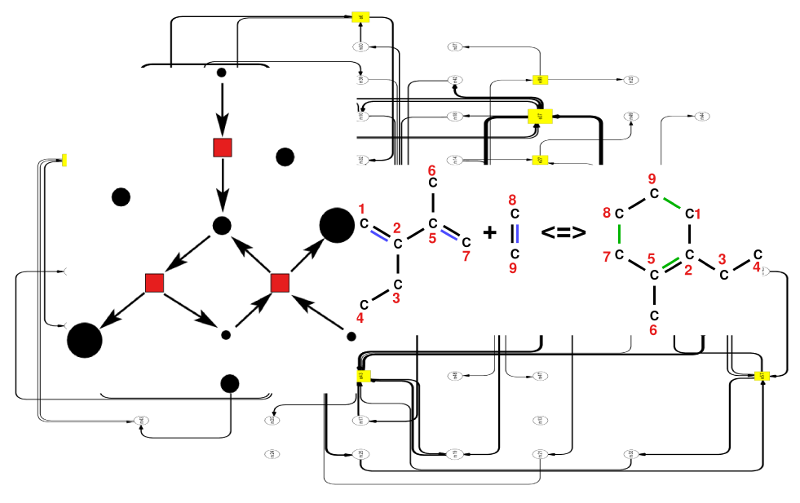
Modeling chemical reaction networks allows us to analyze all possible
reactions within a system and their interconnection via
different substrates.
We developed an efficient graph grammar approach to
describe these changes in connectivity within such networks
over time while cascades of chemical reactions take
place. Using such a formalism permits abstract reasoning about
chemical reaction networks and allows us to identify reaction
mechanisms underlying complex phenomena. We apply our methodology to a
broad set of (bio)chemical problems such as synthesis
planning or the evolution of catalyzed reaction networks.