Index >
Results for CNB 116399-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
116399_ENSG00000082126_HUMAN_1227_1333/1-107 TGCTTCCTCACTCTCAGGGATATTGTCTGGCAGTGGAGGGAGAAGGGGTTCAAAATCTTT
116399_ENSDARG00000008474_ZEBRAFISH_9893_9998/1-107 -GCCTCCTCATTGTCAGACAGATTGTCAGGAAGGGGCGGAAGCACTGGCCCATAATCTCT
116399_ENSMUSG00000026025_MOUSE_1993_2098/1-107 -GCTTCTTCACTGTCGGGGATATTGTCTGGCAGTGGGGGGAGAAGGGGTTCAAAGTCTTT
116399_ENSRNOG00000010486_RAT_9894_9998/1-107 -GCTTCTTCACTGTCGGGGATATTGTCTGGCAGTGGAGGGAGAAGGGGTTCAAAGTCTTT
** ** *** * ** * * ****** ** ** ** ** ** * ** ** * *** *
116399_ENSG00000082126_HUMAN_1227_1333/1-107 CTGAGCTATCGTGTCATGGGCACTGAGCAAGGCCTGGGCACAGGGAA
116399_ENSDARG00000008474_ZEBRAFISH_9893_9998/1-107 CTGAGCCACAGAATCATGGGCAGAGAGCAAAGCCTGTGGAGAAAAAC
116399_ENSMUSG00000026025_MOUSE_1993_2098/1-107 CTGAGCCACTGTGTCATGGGCACTCAGCAAGGCCTAGGGACAGAAGA
116399_ENSRNOG00000010486_RAT_9894_9998/1-107 CTGAGCCACTGTGTCATGGGCCCTCAGCAAAGCCTAGGGACAGAAG-
****** * * ******** ***** **** * * *
116399-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 107
Mean pairwise identity: 78.72
Mean single sequence MFE: -37.25
Consensus MFE: -26.39
Energy contribution: -27.33
Covariance contribution: 0.94
Mean z-score: -1.58
Structure conservation index: 0.71
SVM decision value: 0.16
SVM RNA-class probability: 0.613252
Prediction: RNA
######################################################################
>116399_ENSG00000082126_HUMAN_1227_1333/1-107
UGCUUCCUCACUCUCAGGGAUAUUGUCUGGCAGUGGAGGGAGAAGGGGUUCAAAAUCUUUCUGAGCUAUCGUGUCAUGGGCACUGAGCAAGGCCUGGGCACAGGGAA
..((((((((((.(((((.......))))).))).)))))))....((((((.........))))))...(((((.(((((.((.....))))))))))))...... ( -35.40)
>116399_ENSDARG00000008474_ZEBRAFISH_9893_9998/1-107
GCCUCCUCAUUGUCAGACAGAUUGUCAGGAAGGGGCGGAAGCACUGGCCCAUAAUCUCUCUGAGCCACAGAAUCAUGGGCAGAGAGCAAAGCCUGUGGAGAAAAAC
..((((.((...(((((.(((((((..((..((.((....)).))..)).))))))).)))))(((.((......))))).............)).))))...... ( -35.10)
>116399_ENSMUSG00000026025_MOUSE_1993_2098/1-107
GCUUCUUCACUGUCGGGGAUAUUGUCUGGCAGUGGGGGGAGAAGGGGUUCAAAGUCUUUCUGAGCCACUGUGUCAUGGGCACUCAGCAAGGCCUAGGGACAGAAGA
(((((((((((((((((.......)))))))))))))..(((((((.(....).)))))))))))..((((.((.(((((.((.....))))))))).)))).... ( -39.70)
>116399_ENSRNOG00000010486_RAT_9894_9998/1-107
GCUUCUUCACUGUCGGGGAUAUUGUCUGGCAGUGGAGGGAGAAGGGGUUCAAAGUCUUUCUGAGCCACUGUGUCAUGGGCCCUCAGCAAAGCCUAGGGACAGAAG
(((((((((((((((((.......)))))))))))))..(((((((.(....).)))))))))))..((((.((.(((((..........))))))).))))... ( -38.80)
>consensus
_GCUUCCUCACUGUCAGGGAUAUUGUCUGGCAGUGGAGGGAGAAGGGGUUCAAAAUCUUUCUGAGCCACUGUGUCAUGGGCACUCAGCAAAGCCUAGGGACAGAAAA
..((((((((((((((((.......))))))))))).)))))....((((((.........))))))..(.((((..((((..........))))...)))).)... (-26.39 = -27.33 + 0.94)
116399-rev.rnaz
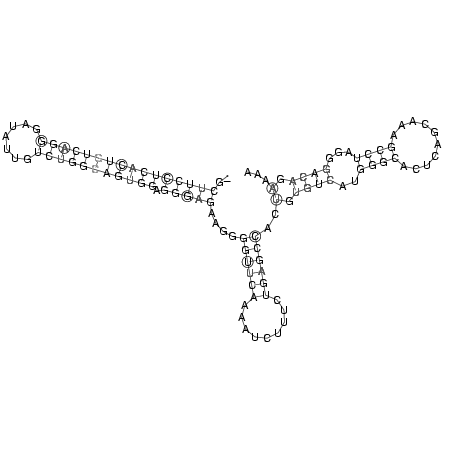
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004