Index >
Results for CNB 231552-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
231552_ENSG00000134107_HUMAN_7051_7118/1-133 GGGAGCGAGTGGGTGGGTTGGAGCTACGTGT----TCTACCCTGTGACTCCAGGCACGTC
231552_ENSRNOG00000007152_RAT_9344_9411/1-133 GGGAGCGAGTGGGTGGGTAGAAGCTACGTGT----TCTACCCTGTGACTCCAAGCACGTC
231552_ENSDARG00000004060_ZEBRAFISH_9664_9767/1-133 GAGTGTATGTGGGTGGATTTGAAGTACGTGTTAAATAAACCCTGTGACTGCAGACACGTC
231552_ENSMUSG00000030103_MOUSE_519_586/1-133 GGGAGCGAGTGGGTGGGTAGAAGCTACGTGT----TCTACCCTGTGACTCCAAGCACGTC
231552_SINFRUG00000133586_FUGU_2671_2800/1-133 -GGAAAGTGGGGGAGAACA-GAGCGACGAGTTAAATAAACCCTCTGACTCCGCGCAGCGC
.*:. .:* ***:*.. : .*. ***:** *.:***** ***** *. .** *
231552_ENSG00000134107_HUMAN_7051_7118/1-133 TGGCC-ACTGCCG-----------------------------------------------
231552_ENSRNOG00000007152_RAT_9344_9411/1-133 TGGCC-GCTGCCG-----------------------------------------------
231552_ENSDARG00000004060_ZEBRAFISH_9664_9767/1-133 TCG-A-GCTCCAGTCACATGAGCCTCACGTGTGTGACGGCGCTTT---------------
231552_ENSMUSG00000030103_MOUSE_519_586/1-133 TGTCC-GCTANNN-----------------------------------------------
231552_SINFRUG00000133586_FUGU_2671_2800/1-133 TGACCTACCTCTGTCACATGAGGCTCACGTGTTAAACGGCGCTGTATCCCCGGCGGAGGG
* . .* .
231552_ENSG00000134107_HUMAN_7051_7118/1-133 -------------
231552_ENSRNOG00000007152_RAT_9344_9411/1-133 -------------
231552_ENSDARG00000004060_ZEBRAFISH_9664_9767/1-133 ------------C
231552_ENSMUSG00000030103_MOUSE_519_586/1-133 -------------
231552_SINFRUG00000133586_FUGU_2671_2800/1-133 ACCGCCTCCTCC-
231552-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 133
Mean pairwise identity: 49.43
Mean single sequence MFE: -32.74
Consensus MFE: -9.26
Energy contribution: -10.78
Covariance contribution: 1.52
Mean z-score: -1.15
Structure conservation index: 0.28
SVM decision value: 0.34
SVM RNA-class probability: 0.696569
Prediction: RNA
######################################################################
>231552_ENSG00000134107_HUMAN_7051_7118/1-133
GGGAGCGAGUGGGUGGGUUGGAGCUACGUGUUCUACCCUGUGACUCCAGGCACGUCUGGCCACUGCCG
..(.((.(((((..((((.(((((.....))))))))).......(((((....))))))))))))). ( -26.10)
>231552_ENSRNOG00000007152_RAT_9344_9411/1-133
GGGAGCGAGUGGGUGGGUAGAAGCUACGUGUUCUACCCUGUGACUCCAAGCACGUCUGGCCGCUGCCG
((.((((.((.((.((((((((.(.....))))))))).(((........)))..)).)))))).)). ( -27.70)
>231552_ENSDARG00000004060_ZEBRAFISH_9664_9767/1-133
GAGUGUAUGUGGGUGGAUUUGAAGUACGUGUUAAAUAAACCCUGUGACUGCAGACACGUCUCGAGCUCCAGUCACAUGAGCCUCACGUGUGUGACGGCGCUUUC
((((.(((((((.(((((((((.(.((((((..........((((....)))))))))))))))).)))).))))))).(((((((....)))).))))))).. ( -36.90)
>231552_ENSMUSG00000030103_MOUSE_519_586/1-133
GGGAGCGAGUGGGUGGGUAGAAGCUACGUGUUCUACCCUGUGACUCCAAGCACGUCUGUCCGCUANNN
...((((..((((.((((((((.(.....))))))))).(((........))).))))..)))).... ( -21.30)
>231552_SINFRUG00000133586_FUGU_2671_2800/1-133
GGAAAGUGGGGGAGAACAGAGCGACGAGUUAAAUAAACCCUCUGACUCCGCGCAGCGCUGACCUACCUCUGUCACAUGAGGCUCACGUGUUAAACGGCGCUGUAUCCCCGGCGGAGGGACCGCCUCCUCC
(((..((.(((((.......(((..((((((...........)))))))))(((((((.......((((........))))....(((.....)))))))))).))))).))(((((.....)))))))) ( -51.70)
>consensus
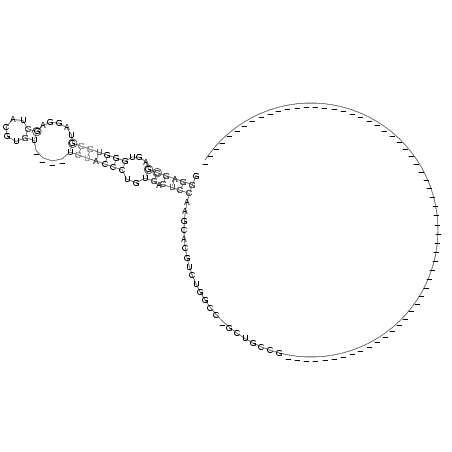
GGGAGCGAGUGGGUGGGUAGGAGCUACGUGU____UCUACCCUGUGACUCCAAGCACGUCUGGCC_GCUGCCG____________________________________________________________
.((((((...(((((((.....((.....))....)))))))..)).)))).................................................................................. ( -9.26 = -10.78 + 1.52)
231552-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004