Index >
Results for CNB 231571
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
231571_ENSG00000134107_HUMAN_6981_7086/1-109 -AGCGTTGTCCAACACGTGAGACTCATGTGA-TGAAGCCGGGGGAGGG-CGGGCAGGTCG
231571_SINFRUG00000142673_FUGU_2286_2356/1-109 AAGCGCCGTCCAACACGTGAGGCTCATGTGA-CGGCGTCGAGTCTGAGTCTGTCCCGGAG
231571_ENSRNOG00000007152_RAT_9274_9378/1-109 -AGCGTTGTCCAACACGTGAGACTCATGTGA-TGAAGCCGGGGGAGGG-CGGGCAGGTCG
231571_ENSDARG00000004060_ZEBRAFISH_9666_9730/1-109 AAGCGCCGTCACACACGTGAGGCTCATGTGACTGGAGCTCGAGACGTGTCTG-CAGTCAC
**** *** ********* ********* * * * * * * *
231571_ENSG00000134107_HUMAN_6981_7086/1-109 CTCCTTCCCTCCCCGGCAGTGGCCAGACGTGCCTGGAGTCACAGGGTAG
231571_SINFRUG00000142673_FUGU_2286_2356/1-109 AGACGTGCCTGC-------------------------------------
231571_ENSRNOG00000007152_RAT_9274_9378/1-109 CTCCTTCCCTCCCCGGCAGCGGCCAGACGTGCTTGGAGTCACAGGGTA-
231571_ENSDARG00000004060_ZEBRAFISH_9666_9730/1-109 AGGGTT-------------------------------------------
*
//
231571.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 109
Mean pairwise identity: 52.13
Mean single sequence MFE: -36.72
Consensus MFE: -15.45
Energy contribution: -16.58
Covariance contribution: 1.13
Mean z-score: -1.26
Structure conservation index: 0.42
SVM decision value: 0.44
SVM RNA-class probability: 0.737380
Prediction: RNA
######################################################################
>231571_ENSG00000134107_HUMAN_6981_7086/1-109
AGCGUUGUCCAACACGUGAGACUCAUGUGAUGAAGCCGGGGGAGGGCGGGCAGGUCGCUCCUUCCCUCCCCGGCAGUGGCCAGACGUGCCUGGAGUCACAGGGUAG
.(((((..(((.((((((.....)))))).....(((((((.((((.(((.((....))))).)))))))))))..)))...))))).((((......)))).... ( -50.60)
>231571_SINFRUG00000142673_FUGU_2286_2356/1-109
AAGCGCCGUCCAACACGUGAGGCUCAUGUGACGGCGUCGAGUCUGAGUCUGUCCCGGAGAGACGUGCCUGC
..((((((((....(((((.....))))))))))))).........((((.((...)).))))........ ( -23.70)
>231571_ENSRNOG00000007152_RAT_9274_9378/1-109
AGCGUUGUCCAACACGUGAGACUCAUGUGAUGAAGCCGGGGGAGGGCGGGCAGGUCGCUCCUUCCCUCCCCGGCAGCGGCCAGACGUGCUUGGAGUCACAGGGUA
.((((.(.....)))))...((((.((((((.((((((((((((((.(((.((....))))).))))))))(((....)))...)).))))...)))))))))). ( -47.60)
>231571_ENSDARG00000004060_ZEBRAFISH_9666_9730/1-109
AAGCGCCGUCACACACGUGAGGCUCAUGUGACUGGAGCUCGAGACGUGUCUGCAGUCACAGGGUU
..(.(((.((((....))))))).).((((((((.(((.((...)).).)).))))))))..... ( -25.00)
>consensus
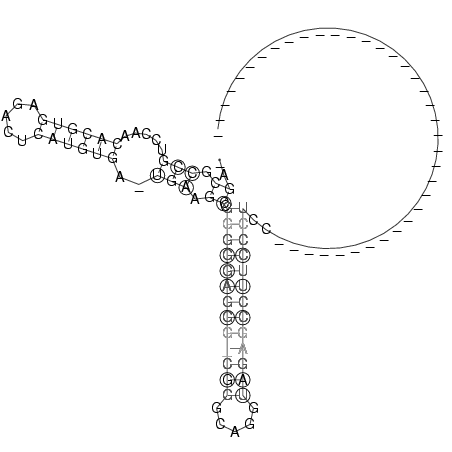
_AGCGCCGUCCAACACGUGAGACUCAUGUGA_UGAAGCCGGGGGAGGG_CGGGCAGGUAGAGCCUUCCCUCC_____________________________________
..((.(((.....((((((.....))))))..))).)).(((((((((((((.....)))))))))))))....................................... (-15.45 = -16.58 + 1.13)
231571.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004