Index >
Results for CNB 260572-rev
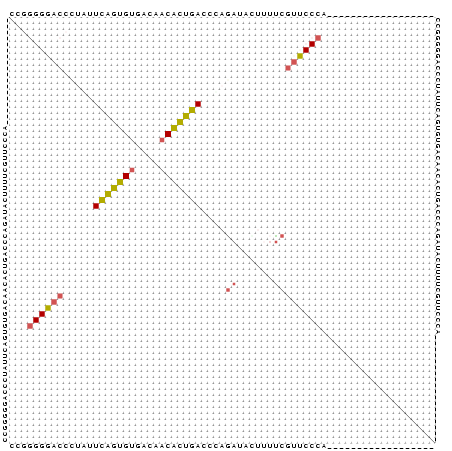
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
260572_ENSG00000154945_HUMAN_9688_9755/1-71 CCGGGGGACCCTGTTCAGTGTGACAACGCTGACCCAGATATTTTCCGTTCCGACCCGGGG
260572_ENSRNOG00000002935_RAT_4048_4115/1-71 CCGGGGGACCCTATTCAGTGTGACAACATTGACCCAGATACTTTTCGTTCCCANNNNNNN
260572_ENSMUSG00000020864_MOUSE_6038_6105/1-71 CCGGGGGACCCTATTCGGTGTGACAACATTGACCTAGATACTTTTCGTTCCCANNNNNNN
260572_SINFRUG00000129036_FUGU_2040_2107/1-71 -CAGGGGACATCTTTTGGTGTGACACCACTGAGCGAAACATTCGT-TCCCCCCCCCCGAT
* ****** ** ******** * *** * * * * * **
260572_ENSG00000154945_HUMAN_9688_9755/1-71 CTCCGGC---G
260572_ENSRNOG00000002935_RAT_4048_4115/1-71 NNNNNNN---N
260572_ENSMUSG00000020864_MOUSE_6038_6105/1-71 NNNNNNN---N
260572_SINFRUG00000129036_FUGU_2040_2107/1-71 ATCCGACTGC-
260572-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 71
Mean pairwise identity: 62.35
Mean single sequence MFE: -19.08
Consensus MFE: -10.96
Energy contribution: -11.40
Covariance contribution: 0.44
Mean z-score: -1.75
Structure conservation index: 0.57
SVM decision value: 1.97
SVM RNA-class probability: 0.984341
Prediction: RNA
######################################################################
>260572_ENSG00000154945_HUMAN_9688_9755/1-71
CCGGGGGACCCUGUUCAGUGUGACAACGCUGACCCAGAUAUUUUCCGUUCCGACCCGGGGCUCCGGCG
((((((((..(((.(((((((....)))))))..))).....))))((((((...)))))).)))).. ( -25.30)
>260572_ENSRNOG00000002935_RAT_4048_4115/1-71
CCGGGGGACCCUAUUCAGUGUGACAACAUUGACCCAGAUACUUUUCGUUCCCANNNNNNNNNNNNNNN
...((((((.....(((((((....)))))))....((......))))))))................ ( -16.60)
>260572_ENSMUSG00000020864_MOUSE_6038_6105/1-71
CCGGGGGACCCUAUUCGGUGUGACAACAUUGACCUAGAUACUUUUCGUUCCCANNNNNNNNNNNNNNN
...((((((.....(((((((....)))))))....((......))))))))................ ( -15.80)
>260572_SINFRUG00000129036_FUGU_2040_2107/1-71
CAGGGGACAUCUUUUGGUGUGACACCACUGAGCGAAACAUUCGUUCCCCCCCCCCGAUAUCCGACUGC
..(((((((((....))))).........((((((.....)))))).....))))............. ( -18.60)
>consensus
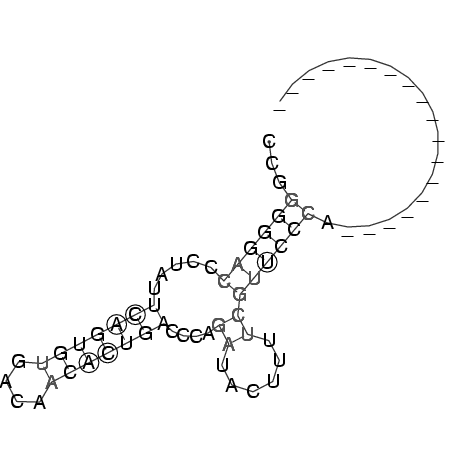
CCGGGGGACCCUAUUCAGUGUGACAACACUGACCCAGAUACUUUUCGUUCCCA__________________
...((((((.....(((((((....)))))))....((......))))))))................... (-10.96 = -11.40 + 0.44)
260572-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004