Index >
Results for CNB 292681
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
292681_ENSG00000181200_HUMAN_9325_9452/1-129 CAGGGCTTTAGTTCAGCAGATGAGCTGCTTGGCTATATTCATACATCTGCTTGCCTGCTG
292681_ENSMUSG00000045383_MOUSE_9441_9561/1-129 ------TTTAGCCCAGCAGATGGGCTGCC-GGCTATATTCATACATCTGCTTGCCTGCTG
292681_ENSDARG00000013366_ZEBRAFISH_9455_9570/1-129 ------TTGGCTTCAGCAGGTGGGCCGCCTGCTTGTGTTCCTACATCTGCCTGCCTGGTC
292681_ENSRNOG00000006531_RAT_9436_9560/1-129 -AGGGCTTGAGCCCTGCAGATGGGCTGCT-GGCTATATTCATACATCTGCTTGCCTGCTG
** * **** ** ** ** * * * *** ********* ****** *
292681_ENSG00000181200_HUMAN_9325_9452/1-129 TGGCAGCTGCTGCTGCTTCACGTTGTGTTCTTTTGTT-CTGTTTCATCCAAGAATGGAAC
292681_ENSMUSG00000045383_MOUSE_9441_9561/1-129 TGGCAGCTGCTGCTGCTTCACGTTGTGTCCTTTTGTT-CTGTTCCATCCAAGAACGGAAT
292681_ENSDARG00000013366_ZEBRAFISH_9455_9570/1-129 TGGCAGCTGCTGTTGTTTAACAACCACTGCTTCCACTGCTGCCTCACTCCAAACTCCTTT
292681_ENSRNOG00000006531_RAT_9436_9560/1-129 TGGCAGCTGCTGCCGCCTCACGTTGTGTCCTTTTGTT-CTGTTCCGTCCAAGAACGGAAT
************ * * ** * *** * *** * * * *
292681_ENSG00000181200_HUMAN_9325_9452/1-129 GCTAGGAAG
292681_ENSMUSG00000045383_MOUSE_9441_9561/1-129 GTGAGGAAG
292681_ENSDARG00000013366_ZEBRAFISH_9455_9570/1-129 GT-------
292681_ENSRNOG00000006531_RAT_9436_9560/1-129 GCGGGGAA-
*
//
292681.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 129
Mean pairwise identity: 71.75
Mean single sequence MFE: -40.53
Consensus MFE: -24.35
Energy contribution: -24.47
Covariance contribution: 0.12
Mean z-score: -1.48
Structure conservation index: 0.60
SVM decision value: 0.06
SVM RNA-class probability: 0.564901
Prediction: RNA
######################################################################
>292681_ENSG00000181200_HUMAN_9325_9452/1-129
CAGGGCUUUAGUUCAGCAGAUGAGCUGCUUGGCUAUAUUCAUACAUCUGCUUGCCUGCUGUGGCAGCUGCUGCUGCUUCACGUUGUGUUCUUUUGUUCUGUUUCAUCCAAGAAUGGAACGCUAGGAAG
((.(((....((..((((((((((((....)))).........)))))))).))..))).))(((((....)))))(((.(...(((((((...(((((..........))))))))))))..)))). ( -39.30)
>292681_ENSMUSG00000045383_MOUSE_9441_9561/1-129
UUUAGCCCAGCAGAUGGGCUGCCGGCUAUAUUCAUACAUCUGCUUGCCUGCUGUGGCAGCUGCUGCUGCUUCACGUUGUGUCCUUUUGUUCUGUUCCAUCCAAGAACGGAAUGUGAGGAAG
...((((((.....))))))((.(((...................))).))((.((((((....)))))).)).......(((((.((((((((((.......)))))))))).))))).. ( -42.81)
>292681_ENSDARG00000013366_ZEBRAFISH_9455_9570/1-129
UUGGCUUCAGCAGGUGGGCCGCCUGCUUGUGUUCCUACAUCUGCCUGCCUGGUCUGGCAGCUGCUGUUGUUUAACAACCACUGCUUCCACUGCUGCCUCACUCCAAACUCCUUUGU
..(((..(((((((..(((.(.......((((....))))).)))..))))...(((.(((.(..((((.....))))..).))).))))))..)))................... ( -33.41)
>292681_ENSRNOG00000006531_RAT_9436_9560/1-129
AGGGCUUGAGCCCUGCAGAUGGGCUGCUGGCUAUAUUCAUACAUCUGCUUGCCUGCUGUGGCAGCUGCUGCCGCCUCACGUUGUGUCCUUUUGUUCUGUUCCGUCCAAGAACGGAAUGCGGGGAA
(((((....)))))((((((((((.....))).........)))))))...(((((.(((((((...)))))))..................(((((((((.......))))))))))))))... ( -46.60)
>consensus
______UUGAGCCCAGCAGAUGGGCUGCC_GGCUAUAUUCAUACAUCUGCUUGCCUGCUGUGGCAGCUGCUGCUGCUUCACGUUGUGUCCUUUUGUU_CUGUUCCAUCCAAGAACGGAAUGCGAGGAA_
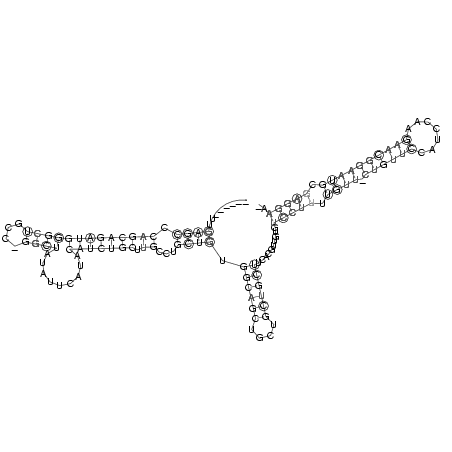
........((((.(((((((((((((....)))).........))))))).))...)))).((((((....))))))..........(((((.((((.((((((.......)))))))))).))))).. (-24.35 = -24.47 + 0.12)
292681.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004