Index >
Results for CNB 294591
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
294591_ENSG00000127946_HUMAN_4733_4795/1-84 T------------------CTCTGAGATACAGA-AATGAAT-TGAGTGACATGAGCAGGA
294591_ENSRNOG00000001448_RAT_6554_6618/1-84 C------------------TTCTGGTCTGTGGTCAACCGAC-TGCCTCTCTCCAGCAACG
294591_ENSMUSG00000039959_MOUSE_6943_7007/1-84 C------------------TTTTGGTCTGTGGTCAACCGAC-TGCCTCTCTCCAGCAACG
294591_ENSDARG00000012291_ZEBRAFISH_7984_8065/1-84 -TTTTCTTTCTCTTTTCTTTCTTCTTCCACAGGCGGTGAGCATCAATAAAGCCATCAACA
294591_SINFRUG00000139764_FUGU_8576_8640/1-84 C------------------TTCTGGGCGGCCGTCACCCGAC-TTCCTCTGTCCAGCAACG
* . . * . .. * . * : * **. .
294591_ENSG00000127946_HUMAN_4733_4795/1-84 TGTGGGTGAGTTTGGAGATGTAC-
294591_ENSRNOG00000001448_RAT_6554_6618/1-84 CCATGCTGTGCTGGAAGTTCTGCC
294591_ENSMUSG00000039959_MOUSE_6943_7007/1-84 CCATGCTGTGCTGGAAGTTTTGTC
294591_ENSDARG00000012291_ZEBRAFISH_7984_8065/1-84 CACAGGAG-GTTGCTGTCAAAGAG
294591_SINFRUG00000139764_FUGU_8576_8640/1-84 CTGTGCTCTGCTGGAAATTCTGCC
* : * * . : :.
//
294591.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 84
Mean pairwise identity: 46.28
Mean single sequence MFE: -16.14
Consensus MFE: -4.46
Energy contribution: -4.26
Covariance contribution: -0.20
Mean z-score: -0.62
Structure conservation index: 0.28
SVM decision value: 0.56
SVM RNA-class probability: 0.780280
Prediction: RNA
######################################################################
>294591_ENSG00000127946_HUMAN_4733_4795/1-84
UCUCUGAGAUACAGAAAUGAAUUGAGUGACAUGAGCAGGAUGUGGGUGAGUUUGGAGAUGUAC
((((..((...((....))..........(((..((.....))..)))..))..))))..... ( -10.90)
>294591_ENSRNOG00000001448_RAT_6554_6618/1-84
CUUCUGGUCUGUGGUCAACCGACUGCCUCUCUCCAGCAACGCCAUGCUGUGCUGGAAGUUCUGCC
((((..((..(..(((....)))..).......(((((......))))).))..))))....... ( -19.10)
>294591_ENSMUSG00000039959_MOUSE_6943_7007/1-84
CUUUUGGUCUGUGGUCAACCGACUGCCUCUCUCCAGCAACGCCAUGCUGUGCUGGAAGUUUUGUC
..........(..(((....)))..).....(((((((..((...))..)))))))......... ( -17.40)
>294591_ENSDARG00000012291_ZEBRAFISH_7984_8065/1-84
UUUUCUUUCUCUUUUCUUUCUUCUUCCACAGGCGGUGAGCAUCAAUAAAGCCAUCAACACACAGGAGGUUGCUGUCAAAGAG
........((((((........(((((...(..((((.((.........))))))..).....)))))........)))))) ( -13.49)
>294591_SINFRUG00000139764_FUGU_8576_8640/1-84
CUUCUGGGCGGCCGUCACCCGACUUCCUCUGUCCAGCAACGCUGUGCUCUGCUGGAAAUUCUGCC
......(((((..(((....)))........(((((((..((...))..)))))))....))))) ( -19.80)
>consensus
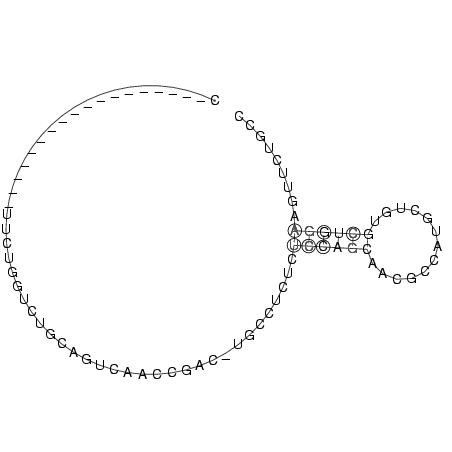
C__________________UUCUGGUCUGCAGUCAACCGAC_UGCCUCUCUCCAGCAACGCCAUGCUGUGCUGGAAGUUCUGCC
..................................................((((((.............))))))......... ( -4.46 = -4.26 + -0.20)
294591.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004