Index >
Results for CNB 391446-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
391446_ENSG00000104529_HUMAN_1690_1757/1-70 -GTCAAACAGGGCCTCGTAGAAGCCCCTCTCGGCTGCATCATACCGGGGCTTCTCCAGCC
391446_ENSMUSG00000022573_MOUSE_181_248/1-70 A-TCAAACAGGGCCTCATAGAAGCCCCTCTCAGCGGCATCGTACCGGGGCTTCTCCAGCC
391446_ENSRNOG00000008084_RAT_182_249/1-70 A-TCAAATAGGGCCTCATAGAAGCCCCTTTCAGCTGCATCATAACGGGGCTTTTCCAGCC
391446_SINFRUG00000136056_FUGU_7740_7804/1-70 G-TTGGCT---CCCTCGTAAAAGCGTTTTTCTGCTTCATCAAAGCGGTGTTTATCGAACC
* **** ** **** * ** ** **** * *** * ** ** * **
391446_ENSG00000104529_HUMAN_1690_1757/1-70 ACACCTCC-C
391446_ENSMUSG00000022573_MOUSE_181_248/1-70 ATACCTCC-C
391446_ENSRNOG00000008084_RAT_182_249/1-70 ATACCTCC-C
391446_SINFRUG00000136056_FUGU_7740_7804/1-70 ATACGTTCT-
* ** * *
391446-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 70
Mean pairwise identity: 73.43
Mean single sequence MFE: -17.07
Consensus MFE: -12.06
Energy contribution: -11.99
Covariance contribution: -0.06
Mean z-score: -1.62
Structure conservation index: 0.71
SVM decision value: 0.95
SVM RNA-class probability: 0.888100
Prediction: RNA
######################################################################
>391446_ENSG00000104529_HUMAN_1690_1757/1-70
GUCAAACAGGGCCUCGUAGAAGCCCCUCUCGGCUGCAUCAUACCGGGGCUUCUCCAGCCACACCUCCC
.........(((...(.(((((((((....((..........))))))))))))..)))......... ( -19.80)
>391446_ENSMUSG00000022573_MOUSE_181_248/1-70
AUCAAACAGGGCCUCAUAGAAGCCCCUCUCAGCGGCAUCGUACCGGGGCUUCUCCAGCCAUACCUCCC
.........(((.....(((((((((.....(((....)))...)))))))))...)))......... ( -21.70)
>391446_ENSRNOG00000008084_RAT_182_249/1-70
AUCAAAUAGGGCCUCAUAGAAGCCCCUUUCAGCUGCAUCAUAACGGGGCUUUUCCAGCCAUACCUCCC
.........(((.....(((((((((..................)))))))))...)))......... ( -16.87)
>391446_SINFRUG00000136056_FUGU_7740_7804/1-70
GUUGGCUCCCUCGUAAAAGCGUUUUUCUGCUUCAUCAAAGCGGUGUUUAUCGAACCAUACGUUCU
(((((.((...(((....))).....((((((.....))))))........)).))).))..... ( -9.90)
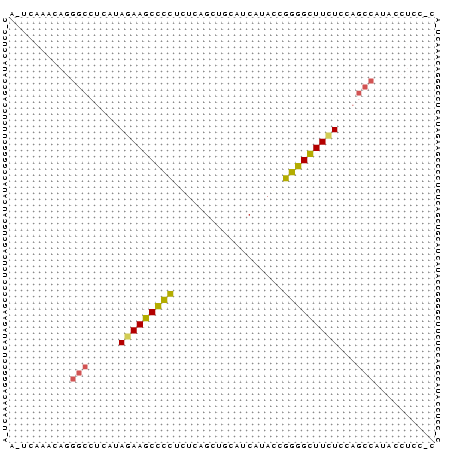
>consensus
A_UCAAACAGGGCCUCAUAGAAGCCCCUCUCAGCUGCAUCAUACCGGGGCUUCUCCAGCCAUACCUCC_C
..........(((.....(((((((((..................)))))))))...))).......... (-12.06 = -11.99 + -0.06)
391446-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004