Index >
Results for CNB 393756-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
393756_ENSG00000138449_HUMAN_8846_8910/1-66 -TTTTTCCTTCTTTCCAAACTTAGCTAACACTGTAGCTGAAGTTGGAAAGGCAAAGCCTT
393756_ENSMUSG00000025993_MOUSE_9749_9813/1-66 -TTTTGTCTTCTTTCCAAACTTAGCTAACACTGTAGCTGAAGTTGGAAAGCCAAAGCCTT
393756_ENSDARG00000000241_ZEBRAFISH_9806_9871/1-66 CCCTTCTCTCCTCTCCAAACTTAGCTATCACTGTAGCTGAAGTCGGAGAGAAATAACCTT
393756_SINFRUG00000122977_FUGU_3355_3417/1-66 -GTTTTCCTCCCCTCCAAACTTAGCTAACACTGTAGCTGAAGTTGGAAAGAAAT-GCTTT
393756_ENSRNOG00000003872_RAT_9698_9762/1-66 -TTATGTCTTCTTTCCAAACTTAGCTAACACTGTAGCTGAAGTTGGAAAGCTAAAGCCTT
* ** * ************** *************** *** ** * * **
393756_ENSG00000138449_HUMAN_8846_8910/1-66 ATGGAA
393756_ENSMUSG00000025993_MOUSE_9749_9813/1-66 ATGGGA
393756_ENSDARG00000000241_ZEBRAFISH_9806_9871/1-66 TCTAGA
393756_SINFRUG00000122977_FUGU_3355_3417/1-66 TATAG-
393756_ENSRNOG00000003872_RAT_9698_9762/1-66 ATGGAA
393756-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 66
Mean pairwise identity: 80.12
Mean single sequence MFE: -18.36
Consensus MFE: -17.20
Energy contribution: -17.56
Covariance contribution: 0.36
Mean z-score: -3.39
Structure conservation index: 0.94
SVM decision value: 3.24
SVM RNA-class probability: 0.998829
Prediction: RNA
######################################################################
>393756_ENSG00000138449_HUMAN_8846_8910/1-66
UUUUUCCUUCUUUCCAAACUUAGCUAACACUGUAGCUGAAGUUGGAAAGGCAAAGCCUUAUGGAA
...((((.....(((((..(((((((......)))))))..)))))(((((...)))))..)))) ( -21.30)
>393756_ENSMUSG00000025993_MOUSE_9749_9813/1-66
UUUUGUCUUCUUUCCAAACUUAGCUAACACUGUAGCUGAAGUUGGAAAGCCAAAGCCUUAUGGGA
(((((....((((((((..(((((((......)))))))..)))))))).)))))((....)).. ( -19.70)
>393756_ENSDARG00000000241_ZEBRAFISH_9806_9871/1-66
CCCUUCUCUCCUCUCCAAACUUAGCUAUCACUGUAGCUGAAGUCGGAGAGAAAUAACCUUUCUAGA
..........((((((.(..((((((((....))))))))..).))))))................ ( -18.20)
>393756_SINFRUG00000122977_FUGU_3355_3417/1-66
GUUUUCCUCCCCUCCAAACUUAGCUAACACUGUAGCUGAAGUUGGAAAGAAAUGCUUUUAUAG
............(((((..(((((((......)))))))..)))))................. ( -13.80)
>393756_ENSRNOG00000003872_RAT_9698_9762/1-66
UUAUGUCUUCUUUCCAAACUUAGCUAACACUGUAGCUGAAGUUGGAAAGCUAAAGCCUUAUGGAA
.........((((((((..(((((((......)))))))..))))))))......((....)).. ( -18.80)
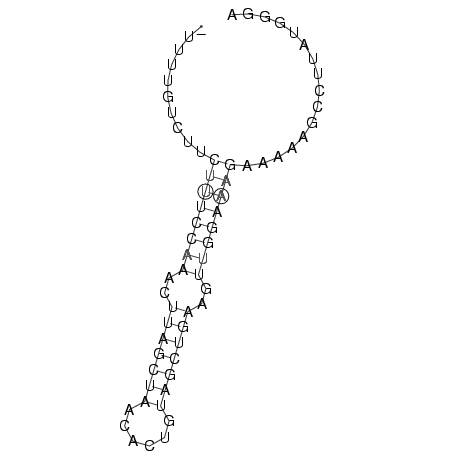
>consensus
_UUUUGUCUUCUUUCCAAACUUAGCUAACACUGUAGCUGAAGUUGGAAAGAAAAAGCCUUAUGGGA
..........((((((((..(((((((......)))))))..))))))))................ (-17.20 = -17.56 + 0.36)
393756-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004