Index >
Results for CNB 456164-rev
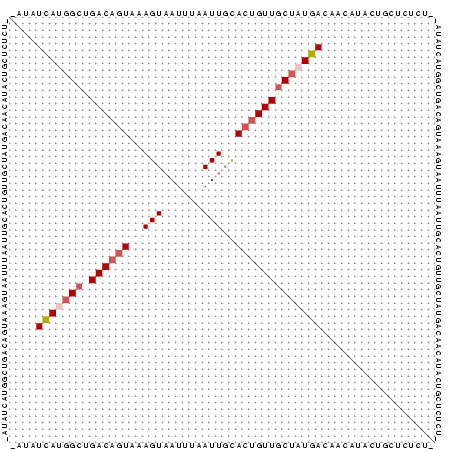
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
456164_ENSG00000053254_HUMAN_2206_2269/1-64 TATATCATGGCTGACAGTAAAGTAATTTAATTGCACTGTTGCTATGACAACATGCTGCTC
456164_SINFRUG00000137365_FUGU_7988_8049/1-64 -ACATTACGGCTGACAGTAAAGTCATCTCATTTTACTGTTGCTATGACAACAAACTCCTC
456164_ENSMUSG00000033713_MOUSE_7447_7508/1-64 -ATATCATGGCTGACAGTAAAGTAATTTAATTGCACTGTTGCTATGACAACATACTGCTC
456164_ENSRNOG00000004709_RAT_7742_7803/1-64 -ATATCATGGCTGACAGTAAAGTAATTTAATTGCACTGTTGCTATGACAACATACTGCTC
456164_ENSDARG00000012833_ZEBRAFISH_1704_1766/1-64 -ATATTACAGCTGACAATAAAGTAATTTAATTGCACAGTTACCATGACAACGGCGGGCTC
* ** * ******* ****** ** * *** ** *** * ******** ***
456164_ENSG00000053254_HUMAN_2206_2269/1-64 TCTT
456164_SINFRUG00000137365_FUGU_7988_8049/1-64 ACT-
456164_ENSMUSG00000033713_MOUSE_7447_7508/1-64 TCT-
456164_ENSRNOG00000004709_RAT_7742_7803/1-64 TCT-
456164_ENSDARG00000012833_ZEBRAFISH_1704_1766/1-64 TGTT
*
456164-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 64
Mean pairwise identity: 83.52
Mean single sequence MFE: -13.66
Consensus MFE: -9.82
Energy contribution: -10.78
Covariance contribution: 0.96
Mean z-score: -2.27
Structure conservation index: 0.72
SVM decision value: 1.14
SVM RNA-class probability: 0.921100
Prediction: RNA
######################################################################
>456164_ENSG00000053254_HUMAN_2206_2269/1-64
UAUAUCAUGGCUGACAGUAAAGUAAUUUAAUUGCACUGUUGCUAUGACAACAUGCUGCUCUCUU
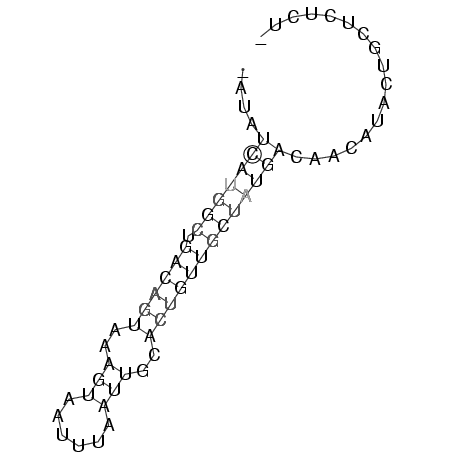
....(((((((.((((((...(((((...))))))))))))))))))................. ( -15.50)
>456164_SINFRUG00000137365_FUGU_7988_8049/1-64
ACAUUACGGCUGACAGUAAAGUCAUCUCAUUUUACUGUUGCUAUGACAACAAACUCCUCACU
...(((..((.(((((((((((......)))))))))))))..)))................ ( -14.20)
>456164_ENSMUSG00000033713_MOUSE_7447_7508/1-64
AUAUCAUGGCUGACAGUAAAGUAAUUUAAUUGCACUGUUGCUAUGACAACAUACUGCUCUCU
...(((((((.((((((...(((((...))))))))))))))))))................ ( -15.50)
>456164_ENSRNOG00000004709_RAT_7742_7803/1-64
AUAUCAUGGCUGACAGUAAAGUAAUUUAAUUGCACUGUUGCUAUGACAACAUACUGCUCUCU
...(((((((.((((((...(((((...))))))))))))))))))................ ( -15.50)
>456164_ENSDARG00000012833_ZEBRAFISH_1704_1766/1-64
AUAUUACAGCUGACAAUAAAGUAAUUUAAUUGCACAGUUACCAUGACAACGGCGGGCUCUGUU
.......(((((.(((((((....))).))))..)))))........(((((......))))) ( -7.60)
>consensus
_AUAUCAUGGCUGACAGUAAAGUAAUUUAAUUGCACUGUUGCUAUGACAACAUACUGCUCUCU_
....(((((((.((((((..(((......)))..)))))))))))))................. ( -9.82 = -10.78 + 0.96)
456164-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004