Index >
Results for CNB 469885
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
469885_ENSG00000169840_HUMAN_9566_9739/1-175 A-GGGAGAGGAGGGGGCGAGGGCGCCTGTTACGGCGTGGGTGGGGTTGACAAGAATAGAG
469885_SINFRUG00000149945_FUGU_9675_9844/1-175 A-AAGAGAGAATGGGCAGTGC-CGCCTCTTGCGGCGTGGGTGGGGCTGACAAGAATAGAC
469885_ENSRNOG00000000952_RAT_9566_9739/1-175 -GGGAAGAGGAGGGGGCGAGTGCGCCTGTTACGGCGTGGGTGGGGTTGACAAGAATAGAG
469885_ENSDARG00000026734_ZEBRAFISH_9427_9598/1-175 A-AAGGGAGAAGGGGCAGTAC-CGCCTCTTGCAGTGTGGGTGGGGCTGACAAGAATAGAC
*** * *** * ***** ** * * ********** *************
469885_ENSG00000169840_HUMAN_9566_9739/1-175 TACATTATGCAGTTCATTTAGTTAACAAGTGAAATAATGAGGAAGCGTGCAGAGTGAATG
469885_SINFRUG00000149945_FUGU_9675_9844/1-175 TACATTATGCAGTTCATTTAGTTAACAAGTGAAATAATGGGGAAGCGTGCAGTGGGAATG
469885_ENSRNOG00000000952_RAT_9566_9739/1-175 TACATTATGCAGTTCATTTAGTTAACAAGTGAAATAATGAGGAAGCGTGCAGAGTGAATG
469885_ENSDARG00000026734_ZEBRAFISH_9427_9598/1-175 TACATTATGCAGTTCATTTAGTTAACAAGTGAAATAATAGGGAAGCGTGCAGAGTGAATG
************************************** ************ * *****
469885_ENSG00000169840_HUMAN_9566_9739/1-175 CCAAGAGAAAAGGCACAAAAACCCTATTGAGAGGCCCCGGCCGCTCCTGGCGTAG
469885_SINFRUG00000149945_FUGU_9675_9844/1-175 CCGAGAGAAA-GGCACAAAA-CCCTATTGAGAGCTTTTGGCCGCTTACAGCGTA-
469885_ENSRNOG00000000952_RAT_9566_9739/1-175 CCAAGAGAAAGGGCACAAAAACCCTATTGAGAGGCCCCGGCCGCTCCTGGCGTAG
469885_ENSDARG00000026734_ZEBRAFISH_9427_9598/1-175 CCGAGAGAAAAGGCACAAAA-CCCTATTGAGAGCTCTCAGCCGCTAGTAGCGTAA
** ******* ********* ************ ****** *****
//
469885.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 175
Mean pairwise identity: 86.41
Mean single sequence MFE: -52.27
Consensus MFE: -34.35
Energy contribution: -37.85
Covariance contribution: 3.50
Mean z-score: -1.55
Structure conservation index: 0.66
SVM decision value: -0.05
SVM RNA-class probability: 0.506078
Prediction: RNA
######################################################################
>469885_ENSG00000169840_HUMAN_9566_9739/1-175
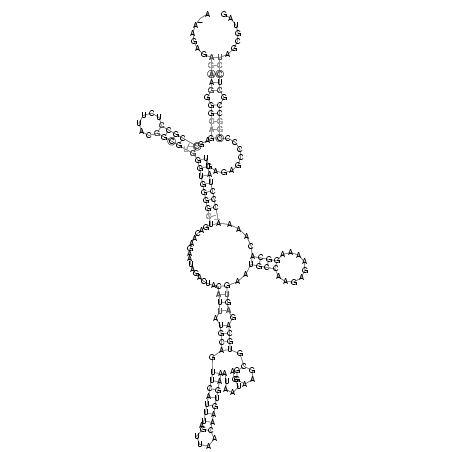
AGGGAGAGGAGGGGGCGAGGGCGCCUGUUACGGCGUGGGUGGGGUUGACAAGAAUAGAGUACAUUAUGCAGUUCAUUUAGUUAACAAGUGAAAUAAUGAGGAAGCGUGCAGAGUGAAUGCCAAGAGAAAAGGCACAAAAACCCUAUUGAGAGGCCCCGGCCGCUCCUGGCGUAG
.(((((.((..(((((....(((((......))))).(((((((((...............((((.((((.(((((((.(....)))))))).......(....).)))).))))..((((.........))))....))))))))).....)))))..)).)))))....... ( -58.10)
>469885_SINFRUG00000149945_FUGU_9675_9844/1-175
AAAGAGAGAAUGGGCAGUGCCGCCUCUUGCGGCGUGGGUGGGGCUGACAAGAAUAGACUACAUUAUGCAGUUCAUUUAGUUAACAAGUGAAAUAAUGGGGAAGCGUGCAGUGGGAAUGCCGAGAGAAAGGCACAAAACCCUAUUGAGAGCUUUUGGCCGCUUACAGCGUA
..........((.((...(((((.....)))))((((((((...................((((((....(((((((.(....)))))))))))))).((((((...(((((((..((((........))))......)))))))...))))))..)))))))).)).)) ( -50.30)
>469885_ENSRNOG00000000952_RAT_9566_9739/1-175
GGGAAGAGGAGGGGGCGAGUGCGCCUGUUACGGCGUGGGUGGGGUUGACAAGAAUAGAGUACAUUAUGCAGUUCAUUUAGUUAACAAGUGAAAUAAUGAGGAAGCGUGCAGAGUGAAUGCCAAGAGAAAGGGCACAAAAACCCUAUUGAGAGGCCCCGGCCGCUCCUGGCGUAG
((((.(.((..(((((....(((((......))))).(((((((((...............((((.((((.(((((((.(....)))))))).......(....).)))).))))..((((.........))))....))))))))).....)))))..)).)))))....... ( -53.70)
>469885_ENSDARG00000026734_ZEBRAFISH_9427_9598/1-175
AAAGGGAGAAGGGGCAGUACCGCCUCUUGCAGUGUGGGUGGGGCUGACAAGAAUAGACUACAUUAUGCAGUUCAUUUAGUUAACAAGUGAAAUAAUAGGGAAGCGUGCAGAGUGAAUGCCGAGAGAAAAGGCACAAAACCCUAUUGAGAGCUCUCAGCCGCUAGUAGCGUAA
...((((((((((((......)))))))(((((((((...(..((....))..)...))))))..)))..(((((((.(....)))))))).((((((((..((.......))...((((.........)))).....))))))))....)))))...(((.....)))... ( -47.00)
>consensus
A_AAGAGAGAAGGGGCAGAGC_CGCCUCUUACGGCGUGGGUGGGGCUGACAAGAAUAGACUACAUUAUGCAGUUCAUUUAGUUAACAAGUGAAAUAAUGAGGAAGCGUGCAGAGUGAAUGCCAAGAGAAAAGGCACAAAA_CCCUAUUGAGAGCCCCCGGCCGCUCCUAGCGUAG
.......(((((.(((((..((((((......))))))(((((((((...............((((.((((.(((((((.(....)))))))).......(....).)))).))))..((((.........))))....))))))))).........))))).)))))....... (-34.35 = -37.85 + 3.50)
469885.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004