BHGbuilder
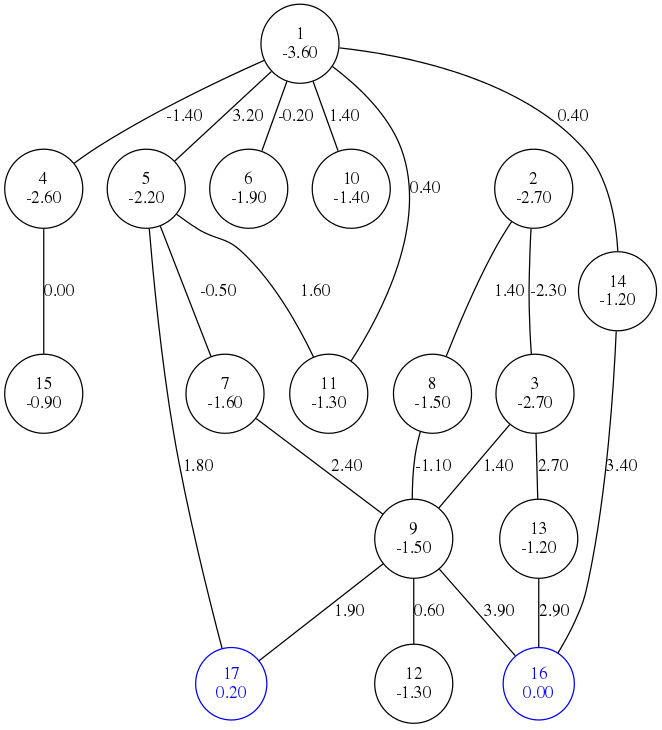 BHGbuilder takes a list of local minima as input and constructs a basin hopping graph abstraction.
Topology is abstracted from paths constructed by findpath procedure from ViennaRNA package.
For every point on path, BHGbuilder searches for local minima by gradient path and stores connections between
neighboring local minima basins. These connections are then being re-evaluated by flooding and printed to output.
There is a multitude of options for interacting with BHG graph to obtain additional data out of graph, such as optimal refolding paths, or refolding rates.
BHGbuilder takes a list of local minima as input and constructs a basin hopping graph abstraction.
Topology is abstracted from paths constructed by findpath procedure from ViennaRNA package.
For every point on path, BHGbuilder searches for local minima by gradient path and stores connections between
neighboring local minima basins. These connections are then being re-evaluated by flooding and printed to output.
There is a multitude of options for interacting with BHG graph to obtain additional data out of graph, such as optimal refolding paths, or refolding rates.
For compiling BHGbuilder, you will need ViennaRNA package installed on your computer (tested with version 2.1.8).
Then follow instructions in README.txt bundled with program. Run ./BHGbuilder -h to get help and available options.
In case you are using our software for your publications you may want to cite:
Basin Hopping Graph: A computational framework to characterize RNA folding landscapes
Marcel Kucharík, Ivo L. Hofacker, Peter F. Stadler and Jing Qin
Bioinformatics 2014
doi:10.1093/bioinformatics/btu156
bibtex

 tbi.univie.ac.at -- programming
tbi.univie.ac.at -- programming