Basin hopping graph
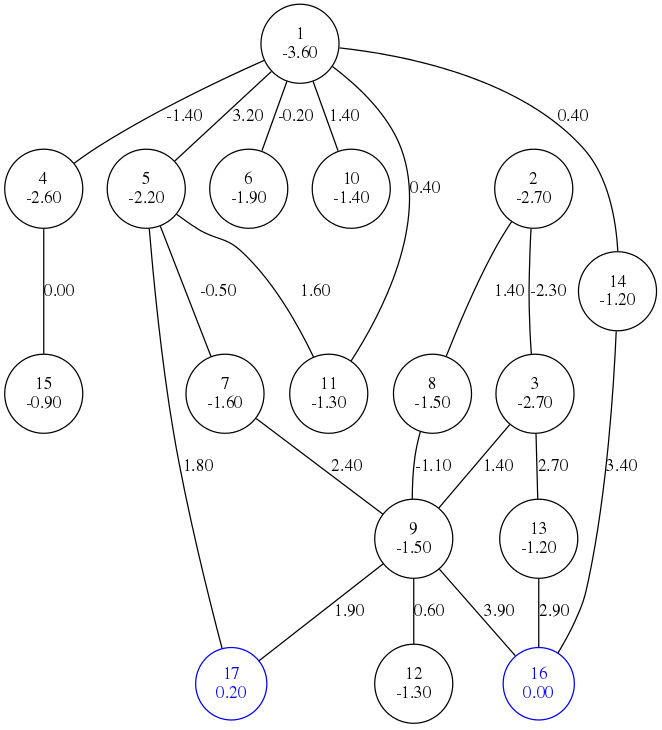 Basin hopping graph framework aims to construct a landscape abstraction, or a basin hopping graph (BHG), of RNA secondary structure landscape of a chosen sequence. Nodes of BHG represent local minima basins and edges available direct connections.
Edge weights correspond to saddle energy of the connection. Framework uses many heuristics, so resulting graph is NOT exact every time. Exact enumeration techniques are unfortunately limited to small RNA molecules.
BHG framework consists of three parts:
Basin hopping graph framework aims to construct a landscape abstraction, or a basin hopping graph (BHG), of RNA secondary structure landscape of a chosen sequence. Nodes of BHG represent local minima basins and edges available direct connections.
Edge weights correspond to saddle energy of the connection. Framework uses many heuristics, so resulting graph is NOT exact every time. Exact enumeration techniques are unfortunately limited to small RNA molecules.
BHG framework consists of three parts:
- local minima extraction routine RNAlocmin
- adaptive search scheme for efficient local minima generation
- basin hopping graph construction program BHGbuilder
RNAlocmin alone is just improved gradient-walking routine for searching a local minima from structures. Adaptive search with RNAlocmin is responsible for generating local minima in an efficient and highly adjustable way.
Standalone program BHGbuilder then takes these local minima (or local minima generated from elsewhere, for example with barriers) and creates a basin hopping graph abstraction of the landscape. Output of BHGbuilder can provide great insight into topology of particular landscape through multitude of available outputs, e.g: saddle heights between basins, actual drawing of graph, optimal refolding paths, or kinetic rates for treekin program.
In case you are using our software for your publications you may want to cite:
Basin Hopping Graph: A computational framework to characterize RNA folding landscapes
Marcel Kucharík, Ivo L. Hofacker, Peter F. Stadler and Jing Qin
Bioinformatics 2014
doi:10.1093/bioinformatics/btu156
bibtex

 tbi.univie.ac.at -- programming
tbi.univie.ac.at -- programming