Neighborhood Relation and Move Sets for Secondary Structures
Different functions to generate structural neighbors of a secondary structure according to a particular Move Set.
This module contains methods to compute the neighbors of an RNA secondary structure. Neighbors of a given structure are all structures that differ in exactly one base pair. That means one can insert an delete base pairs in the given structure. These insertions and deletions of base pairs are usually called moves. A third move which is considered in these methods is a shift move. A shifted base pair has one stable position and one position that changes. These moves are encoded as follows:
insertion: \((i, j)\) where \(i,j > 0\)
deletion: \((i, j)\) where \(i,j\) < 0
shift: \((i, j)\) where either \(i > 0, j < 0\) or \(i < 0, j > 0\)
The negative position of a shift indicates the position that has changed.
An example:
We are given a sequence and a structure.
Sequence AAGGAAACC
Structure ..(.....)
Indices 123456789
The given base pair is (3,9) and the neighbors are the insertion (4, 8),
the deletion (-3,-9), the shift (3,-8) and the shift (-4, 9).
This leads to the neighbored structures:
...(....)
.........
...(...).
....(...)
A simple method to construct all insertions is to iterate over the positions of a sequence twice. The first iteration has the index i in [1, sequence length], the second iteration has the index j in [i+1, sequence length]. All pairs (i,j) with compatible letters and which are non-crossing with present base pairs are valid neighbored insertion moves. Valid deletion moves are all present base pairs with negative sign. Valid shift moves are constructed by taking all paired positions as fix position of a shift move and iterating over all positions of the sequence. If the letters of a position are compatible and if it the move is non-crossing with existing base pairs, we have a valid shift move. The method of generating shift moves can be accelerated by skipping neighbored base pairs.
If we need to construct all neighbors several times for subsequent moves, we can speed up the task by using the move set of the previous structure. The previous move set has to be filtered, such that all moves that would cross the next selected move are non-crossing. Next, the selected move has to be removed. Then one has to only to generate all moves that were not possible before. One move is the inverted selected move (if it was an insertion, simply make the indices negative). The generation of all other new moves is different and depends on the selected move. It is easy for an insertion move, because we have only to include all non-crossing shift moves, that are possible with the new base pair. For that we can either iterate over the sequence or we can select all crossing shift moves in the filter procedure and convert them into shifts.
The generation of new moves given a deletion is a little bit more complex, because we can create more moves. At first we can insert the deleted pair as insertion move. Then we generate all insertions that would have crossed the deleted base pair. Finally we construct all crossing shift moves.
If the given move is a shift, we can save much time by specifying the intervals for the generation of new moves. The interval which was enclosed by the positive position of the shift move and the previous paired position is the freed interval after applying the move. This freed interval includes all positions and base pairs that we need to construct new insertions and shifts. All these new moves have one position in the freed interval and the other position in the environment of the freed interval. The environment are all position which are outside the freed interval, but within the same enclosing loop of the shift move. The environment for valid base pairs can be divided into one or more intervals, depending on the shift move. The following examples describe a few scenarios to specify the intervals of the environment.
Example 1:
increase the freed interval with a shift move
AAAAGACAAGAAACAAAAGAGAAACAACAAACAAGAAACAAACAAAA
....(....(...)....(.(...)..)......(...)...).... // structure before the shift
....(....(...)....(.(...)......)..(...)...).... // structure after the shift
............................[__]............... // freed interval
..................[________]................... // interval that can pair with the freed interval
Example 2:
switch the freed interval with a shift move
AAAAGACAAGAAACAAAAGAGAAACAACAAACAAGAAACAAACAAAA
....(....(...)....(.(...)..)......(...)...).... // structure before the shift
....(.(..(...)....).(...).........(...)...).... // structure after the shift
...................[_______]................... // freed interval
....[_].....................[_____________].... // intervals that can pair with the freed interval
Example 3:
decrease the freed interval with a shift move
AAAAGACAAGAAACAAAAGAGAAACAACAAACAAGAAACAAACAAAA
....(....(...)....(.(...)......)..(...)...).... // structure before the shift
....(....(...)....(.(...)..)......(...)...).... // structure after the shift
............................[__]............... // freed interval
....[____________].............[__________].... // intervals that can pair with the freed interval
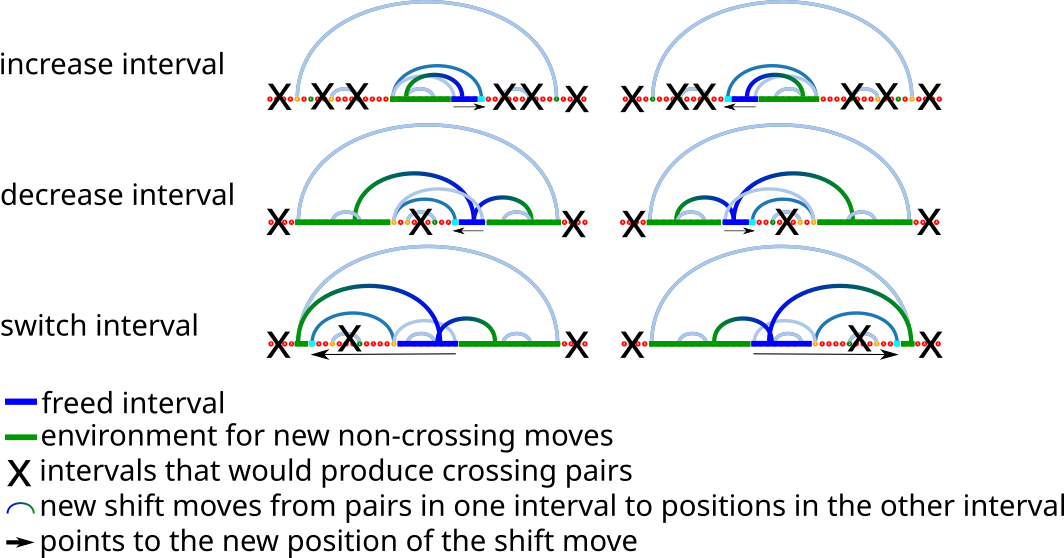
Given the intervals of the environment and the freed interval, the new shift moves can be constructed quickly. One has to take all positions of pairs from the environment in order to create valid pairs with positions in the freed interval. The same procedure can be applied for the other direction. This is taking all paired positions within the freed interval in order to look for pairs with valid positions in the intervals of the environment.
Defines
-
VRNA_MOVESET_INSERTION
- #include <ViennaRNA/landscape/move.h>
Option flag indicating insertion move.
See also
-
VRNA_MOVESET_DELETION
- #include <ViennaRNA/landscape/move.h>
Option flag indicating deletion move.
See also
-
VRNA_MOVESET_SHIFT
- #include <ViennaRNA/landscape/move.h>
Option flag indicating shift move.
See also
-
VRNA_MOVESET_NO_LP
- #include <ViennaRNA/landscape/move.h>
Option flag indicating moves without lonely base pairs.
See also
-
VRNA_MOVESET_DEFAULT
- #include <ViennaRNA/landscape/move.h>
Option flag indicating default move set, i.e. insertions/deletion of a base pair.
See also
-
VRNA_MOVE_NO_APPLY
- #include <ViennaRNA/landscape/move.h>
-
VRNA_NEIGHBOR_CHANGE
- #include <ViennaRNA/landscape/neighbor.h>
State indicator for a neighbor that has been changed.
See also
-
VRNA_NEIGHBOR_INVALID
- #include <ViennaRNA/landscape/neighbor.h>
State indicator for a neighbor that has been invalidated.
See also
-
VRNA_NEIGHBOR_NEW
- #include <ViennaRNA/landscape/neighbor.h>
State indicator for a neighbor that has become newly available.
See also
Typedefs
-
typedef struct vrna_move_s vrna_move_t
- #include <ViennaRNA/landscape/move.h>
A single move that transforms a secondary structure into one of its neighbors.
-
typedef void (*vrna_move_update_f)(vrna_fold_compound_t *fc, vrna_move_t neighbor, unsigned int state, void *data)
- #include <ViennaRNA/landscape/neighbor.h>
Prototype of the neighborhood update callback.
See also
vrna_move_neighbor_diff_cb(), VRNA_NEIGHBOR_CHANGE, VRNA_NEIGHBOR_INVALID, VRNA_NEIGHBOR_NEW
- Param fc:
The fold compound the calling function is working on
- Param neighbor:
The move that generates the (changed or new) neighbor
- Param state:
The state of the neighbor (move) as supplied by argument
neighbor- Param data:
Some arbitrary data pointer as passed to vrna_move_neighbor_diff_cb()
- void() vrna_callback_move_update (vrna_fold_compound_t *fc, vrna_move_t neighbor, unsigned int state, void *data)
- #include <ViennaRNA/landscape/neighbor.h>
Functions
-
vrna_move_t vrna_move_init(int pos_5, int pos_3)
- #include <ViennaRNA/landscape/move.h>
Create an atomic move.
See also
- Parameters:
pos_5 – The 5’ position of the move (positive for insertions, negative for removal, any value for shift moves)
pos_3 – The 3’ position of the move (positive for insertions, negative for removal, any value for shift moves)
- Returns:
An atomic move as specified by
pos_5andpos_3
-
void vrna_move_list_free(vrna_move_t *moves)
- #include <ViennaRNA/landscape/move.h>
delete all moves in a zero terminated list.
-
void vrna_move_apply(short *pt, const vrna_move_t *m)
- #include <ViennaRNA/landscape/move.h>
Apply a particular move / transition to a secondary structure, i.e. transform a structure.
- Parameters:
pt – [inout] The pair table representation of the secondary structure
m – [in] The move to apply
-
void vrna_move_apply_db(char *structure, const short *pt, const vrna_move_t *m)
- #include <ViennaRNA/landscape/move.h>
-
int vrna_move_is_removal(const vrna_move_t *m)
- #include <ViennaRNA/landscape/move.h>
Test whether a move is a base pair removal.
- Parameters:
m – The move to test against
- Returns:
Non-zero if the move is a base pair removal, 0 otherwise
-
int vrna_move_is_insertion(const vrna_move_t *m)
- #include <ViennaRNA/landscape/move.h>
Test whether a move is a base pair insertion.
- Parameters:
m – The move to test against
- Returns:
Non-zero if the move is a base pair insertion, 0 otherwise
-
int vrna_move_is_shift(const vrna_move_t *m)
- #include <ViennaRNA/landscape/move.h>
Test whether a move is a base pair shift.
- Parameters:
m – The move to test against
- Returns:
Non-zero if the move is a base pair shift, 0 otherwise
-
int vrna_move_compare(const vrna_move_t *m, const vrna_move_t *b, const short *pt)
- #include <ViennaRNA/landscape/move.h>
Compare two moves.
The function compares two moves
mandband returns whether movemis lexicographically smaller (-1), larger (1) or equal to moveb.If any of the moves
morbis a shift move, this comparison only makes sense in a structure context. Thus, the third argument with the current structure must be provided.Note
This function returns 0 (equality) upon any error, e.g. missing input
Warning
Currently, shift moves are not supported!
- Parameters:
m – The first move of the comparison
b – The second move of the comparison
pt – The pair table of the current structure that is compatible with both moves (maybe NULL if moves are guaranteed to be no shifts)
- Returns:
-1 if
m<b, 1 ifm>b, 0 otherwise
-
void vrna_loopidx_update(int *loopidx, const short *pt, int length, const vrna_move_t *m)
- #include <ViennaRNA/landscape/neighbor.h>
Alters the loopIndices array that was constructed with vrna_loopidx_from_ptable().
The loopIndex of the current move will be inserted. The correctness of the input will not be checked because the speed should be optimized.
- Parameters:
loopidx – [inout] The loop index data structure that needs an update
pt – [in] A pair table on which the move will be executed
length – The length of the structure
m – [in] The move that is applied to the current structure
-
vrna_move_t *vrna_neighbors(vrna_fold_compound_t *fc, const short *pt, unsigned int options)
- #include <ViennaRNA/landscape/neighbor.h>
Generate neighbors of a secondary structure.
This function allows one to generate all structural neighbors (according to a particular move set) of an RNA secondary structure. The neighborhood is then returned as a list of transitions / moves required to transform the current structure into the actual neighbor.
- SWIG Wrapper Notes:
This function is attached as an overloaded method
neighbors()to objects of typefold_compound. The optional parameteroptionsdefaults to VRNA_MOVESET_DEFAULT if it is omitted. See, e.g.RNA.fold_compound.neighbors()in the Python API.
See also
vrna_neighbors_successive(), vrna_move_apply(), VRNA_MOVESET_INSERTION, VRNA_MOVESET_DELETION, VRNA_MOVESET_SHIFT, VRNA_MOVESET_DEFAULT
- Parameters:
fc – [in] A vrna_fold_compound_t containing the energy parameters and model details
pt – [in] The pair table representation of the structure
options – Options to modify the behavior of this function, e.g. available move set
- Returns:
Neighbors as a list of moves / transitions (the last element in the list has both of its fields set to 0)
-
vrna_move_t *vrna_neighbors_successive(const vrna_fold_compound_t *fc, const vrna_move_t *curr_move, const short *prev_pt, const vrna_move_t *prev_neighbors, int size_prev_neighbors, int *size_neighbors, unsigned int options)
- #include <ViennaRNA/landscape/neighbor.h>
Generate neighbors of a secondary structure (the fast way)
This function implements a fast way to generate all neighbors of a secondary structure that results from successive applications of individual moves. The speed-up results from updating an already known list of valid neighbors before the individual move towards the current structure took place. In essence, this function removes neighbors that are not accessible anymore and inserts neighbors emerging after a move took place.
See also
vrna_neighbors(), vrna_move_apply(), VRNA_MOVESET_INSERTION, VRNA_MOVESET_DELETION, VRNA_MOVESET_SHIFT, VRNA_MOVESET_DEFAULT
- Parameters:
fc – [in] A vrna_fold_compound_t containing the energy parameters and model details
curr_move – [in] The move that was/will be applied to
prev_ptprev_pt – [in] A pair table representation of the structure before
curr_moveis/was appliedprev_neighbors – [in] The list of neighbors of
prev_ptsize_prev_neighbors – The size of
prev_neighbors, i.e. the lists lengthsize_neighbors – [out] A pointer to store the size / length of the new neighbor list
options – Options to modify the behavior of this function, e.g. available move set
- Returns:
Neighbors as a list of moves / transitions (the last element in the list has both of its fields set to 0)
-
int vrna_move_neighbor_diff_cb(vrna_fold_compound_t *fc, short *ptable, vrna_move_t move, vrna_move_update_f cb, void *data, unsigned int options)
- #include <ViennaRNA/landscape/neighbor.h>
Apply a move to a secondary structure and indicate which neighbors have changed consequentially.
This function applies a move to a secondary structure and explores the local neighborhood of the affected loop. Any changes to previously compatible neighbors that have been affected by this loop will be reported through a callback function. In particular, any of the three cases might appear:
A previously available neighbor move has changed, usually the free energy change of the move (VRNA_NEIGHBOR_CHANGE)
A previously available neighbor move became invalid (VRNA_NEIGHBOR_INVALID)
A new neighbor move becomes available (VRNA_NEIGHBOR_NEW)
See also
vrna_move_neighbor_diff(), VRNA_NEIGHBOR_CHANGE, VRNA_NEIGHBOR_INVALID, VRNA_NEIGHBOR_NEW, vrna_move_update_f, #VRNA_MOVE_NO_APPLY
- Parameters:
fc – A fold compound for the RNA sequence(s) that this function operates on
ptable – The current structure as pair table
move – The move to apply
cb – The address of the callback function that is passed the neighborhood changes
data – An arbitrary data pointer that will be passed through to the callback function
cboptions – Options to modify the behavior of this function, .e.g available move set
- Returns:
Non-zero on success, 0 otherwise
-
vrna_move_t *vrna_move_neighbor_diff(vrna_fold_compound_t *fc, short *ptable, vrna_move_t move, vrna_move_t **invalid_moves, unsigned int options)
- #include <ViennaRNA/landscape/neighbor.h>
Apply a move to a secondary structure and indicate which neighbors have changed consequentially.
Similar to vrna_move_neighbor_diff_cb(), this function applies a move to a secondary structure and reports back the neighbors of the current structure become affected by this move. Instead of executing a callback for each of the affected neighbors, this function compiles two lists of neighbor moves, one that is returned and consists of all moves that are novel or may have changed in energy, and a second,
invalid_moves, that consists of all the neighbor moves that become invalid, respectively.- Parameters:
fc – A fold compound for the RNA sequence(s) that this function operates on
ptable – The current structure as pair table
move – The move to apply
invalid_moves – The address of a move list where the function stores those moves that become invalid
options – Options to modify the behavior of this function, .e.g available move set
- Returns:
A list of moves that might have changed in energy or are novel compared to the structure before application of the move
-
struct vrna_move_s
- #include <ViennaRNA/landscape/move.h>
An atomic representation of the transition / move from one structure to its neighbor.
An atomic transition / move may be one of the following:
a base pair insertion,
a base pair removal, or
a base pair shift where an existing base pair changes one of its pairing partner.
These moves are encoded by two integer values that represent the affected 5’ and 3’ nucleotide positions. Furthermore, we use the following convention on the signedness of these encodings:
both values are positive for insertion moves
both values are negative for base pair removals
both values have different signedness for shift moves, where the positive value indicates the nucleotide that stays constant, and the others absolute value is the new pairing partner
Note
A value of 0 in either field is used as list-end indicator and doesn’t represent any valid move.
Public Members
-
int pos_5
The (absolute value of the) 5’ position of a base pair, or any position of a shifted pair.
-
int pos_3
The (absolute value of the) 3’ position of a base pair, or any position of a shifted pair.
-
vrna_move_t *next
The next base pair (if an elementary move changes more than one base pair), or
NULLHas to be terminated with move 0,0.
-
VRNA_MOVESET_INSERTION