Changelog
Version 2.7.x
-
Programs
- Allow for FASTA header changes in
ct2db - Fix constraints parsing in
RNAsubopt - Add new command line options for structure probing data to
RNAfoldandRNAsubopt
- Allow for FASTA header changes in
-
Library
- API: Refactor structure probing data API to support multiple data at once, position-wise weighting, user-defined reactivity-to-energy strategies and more
- API: Refactor hard constraint wrappers
- API: Add
ViennaRNA/math/to provide several useful mathematical functions - API: Add
vrna_str_to_dbl_array() - API: Add
vrna_file_doubles_read() - API: Fix backtracking issues in exterior loop
- API: Fix backtracking issues with odd dangles
- API: Fix segmentation fault in
vrna_pt_pk_remove() - API: Fix
vrna_plist()andvrna_ptable()to deal with malformed input - API: Fix soft constraints access in backtracking of QM2 (Boltzmann sampling)
- SWIG: Add wrappers for concentration dependency computations
- SWIG: Fix
RNA.fold_compound.bpp()output for multistrand base pair probabilities - SWIG: Do not return symmetric pair probability matrix by default
- SWIG: Fix Python object ownership in callbacks for unstructured domains, direct soft constraints and landscape moves
-
Package
- Update
dlibto version 20.0 - Overall improvements to the documentation
- Fix energy parameter file markup
- Fix compilation with compilers that default to C23
- Update
swigrequirement to 4.2.0 - Fix compilation without
libsvm - Update
breatheto version v5.0.0a5 - Add probing data priors (paired and unpaired) for 1M7 and DMS
- Update
This version adds G-Quadruplex support for circular RNAs, a new and unified experiment
RNA structure probing API and an internal log-message system. All new features are exposed either through
the command line tools or(and) the API of RNAlib.
For multibranch loop decompositions, our global RNA structure prediction recursions now rely on an M2 (at least two branches within a multiloop)
dynamic programming matrix, rather the the M1 DP matrix (exactly one branch within a multiloop) that has been used before. This not only slightly
reduces the overall runtime but also unifies the implementations for circular RNAs, global and local RNA structure prediction. Moreover, the API underwent
a major clean-up to unify the naming of functions and the data types of their input and output parameters. RNAlib header include files have
now been grouped into separate subdirectories to obtain a better structured interface.
With this release we also ship two new sets of energy parameters for modified bases, in particular m5C and N1-methyl-pseudouridine. Both parameter sets have been taken from literature and are derived from in-silico computations. Therefore, we did not hard-wire them into the core library just yet, but only provide the parameter files for manually loading them at runtime.
The user- and developer manuals have been heavily worked on and are available as a unified documentation at ReadTheDocs.
The list of all major changes in this version can be found below:
-
Programs
- Add hard limit for number of input structures in
RNAdistance - Add counter example settings to
RNAalifold - Add covariance annotation legend to
RNAplotlayout plots for MSA input - Add covariance annotation legend to
RNALalifoldlayout plots - Adapt structure conservation coloring and add legend in alignment output of
RNAplot,RNAalifold, andRNALalifold - Add
RNAconsensusPython program that will eventually replacerefold.pl - Add
--log-file,--log-level,--log-calland--log-timecommand line options to executable programs - Add
--betaScaleand--pfScaleoptions to and rescale Boltzmann factors inRNAPKplex - Add support for G-Quadruplexes in circular RNAs for
RNAfold,RNAalifold, andRNAeval - Change
RNAplotcommand line argument-oto-f - Add
--random-seedoption toRNAsuboptandRNAalifoldto specify seed for random number generator
- Add hard limit for number of input structures in
-
Library
- API: Add circular RNA G-Quadruplex support
- API: Add structure prediction benchmark functions
vrna_compare_structure()andvrna_compare_structure_pt() - API: Add
vrna_annotate_covar_pt()that allows for specifying number of counter examples - API: Add structure conservation legend to EPS consensus structure layout plots
- API: Add
vrna_string_make_space_for()andvrna_string_available_space()functions inViennaRNA/datastructures/string.h - API: Add
vrna_file_PS_aln_opt()to allow for changing conservation coloring - API: Add flexible log message system to avoid spam on
stderrandstdout - API: Add (generic) compressed sparse row (CSR) matrix implementation
- API: Add Eddy 2014 approach to incorporate experimental probing data (using Gaussian KDE)
- API: Add generic support for experimental probing data via new API in
ViennaRNA/probing/basic.h - API: Add full probing data support for consensus structure prediction
- API: Add
vrna_sc_multi_cb_add_comparative()to allow for multi callback soft constraints in comparative structure predictions - API: Add
vrna_fold_compound_tto parameters passed to recursion status callback - API: Add M2 matrices in favor of M1 for global MFE and partition function computations
- API: Add safeguard to
vrna_array_free() - API: Add
vrna_pairing_tendency()as replacement forvrna_db_from_probs() - API: Group related API symbols and header files into specific subdirectories
- API: Allow base pair hard constraints via commands file where
j = i + 1 - API: Refactor
vrna_pk_plex()accessibility computations - API: Refactor backtracking implementations and API, now located under
ViennaRNA/backtrack/ - API: Refactor experimental probing data (SHAPE) implementations, now located under
ViennaRNA/probing/ - API: Refactor auxiliary grammar extension API, now located under
ViennaRNA/grammar/ - API: Refactor G-Quadruplex implementation and fix existing bugs and numerical issues in corresponding energy evaluation
- API: Refactor verbose
vrna_eval*()implementations - API: Refactor structure plotting API and add unified structure plotting function
vrna_plot_structure() - API: Remove
exit()calls fromRNAlib - API: Change behavior and parameter order for
vrna_hc_add_bp_strand() - API: Change behavior and parameters of
vrna_hc_add_up_strand() - API: Unify backtracking matrix flags
- API: Use
vrna_array()based base pair and backtrack stacks for MFE implementations - API: Introduce entropic penalty for unpaired circular RNAs (may be switched off by model settings
circ_penaltyflag) - API: Deprecate
vrna_message_info(),vrna_message_warning(), andvrna_message_error()in favor of new loggin system - API: Fix
vrna_neighbor_diff*()insertion moves - API: Fix MFE inside recursion for multiloop unpaired positions
- API: Fix
vrna_hx_from_ptable()when provided hairpins of length 0 - API: Fix
vrna_hx_merge()to support pseudoknotted helices - API: Fix re-use of previous energy parameters upon model changes in
duplex.c - API: Fix re-use of previous energy parameters upon model changes in
c_plex.c - API: Fix re-use of previous energy parameters upon model changes in
plex.c - API: Fix re-use of previous energy parameters upon model changes in
ali_plex.c - API: Fix re-use of previous energy parameters upon model changes in
snofold.c - API: Fix re-use of previous energy parameters upon model changes in
snoop.c - API: Fix mismatch and dangling end energies in
sc_cb_mod.cfor modified base support - API: Fix Zuker-style subopt backtracking
- API: Fix bug in
vrna_strdup_vprintf()that resulted in losing parmeters upon consecutive calls - API: Fix default exterior loop MFE soft constraints for comparative structure prediction
- SWIG: Use
av_len()instad ofav_top_index()for Perl 5 to support perl5 < v5.18.0 - SWIG: Fix compilation issues for Python 3.12 due to use of
SWIG_Python_str_AsChar() - SWIG: Add wrapper for
vrna_hx_merge() - SWIG: Add wrapper for
vrna_sc_multi_cb_add() - SWIG: Add wrappers for
vrna_hc_add_bp_strand()andvrna_hc_add_up_strand() - SWIG: Add file I/O constants
- SWIG: Add wrappers for structure prediction benchmark functions
- SWIG: Add wrappers for log message system
- SWIG: Add wrapper for
vrna_stack_prob() - SWIG: Add wrappers for new probing data API
- SWIG: Add wrapper for
vrna_n_multichoose_k() - SWIG: Return
intinstead of float foreval_structure_pt_simple()
-
Package
- AUTOCONF: Fix several
autoconf/automakerelated issues - AUTOCONF: Add
./configure --enable-debugoption that prevents removal ofvrna_log_debug()message fromRNAlib - Add m5C JSON energy parameter file
- Add N1-methylpseudouridine JSON energy parameter file
- Add OpenMP library flags to
RNAlib2.pc - DOC: Improve document structure
- Bump
libsvmto version 3.35 - Remove RNA-Tutorial since it is now included as part of the reference manual
- Remove Python 2 builds from MacOSX installer
- AUTOCONF: Fix several
Version 2.6.x
-
Programs
- Fix C++17 compilation issue with
kinwalker - Fix potential compilation issues with C++20 in
RNAforesterfrontend - Refactor and correct spelling issues in man pages for several executable programs
- Fix C++17 compilation issue with
-
Library
- API: Add shift move support to
vrna_move_neighbor_diff*()functions - API: Fix char array initialization in
snoop.c - API: Fix potentially leaking file pointer in
vrna_file_msa_read() - API: Fix potentially leaking memory in
rnaplot_EPS() - API: Fix potential use of uninitialized variable in
vrna_rotational_symmetry_db_pos() - API: Fix soft constraints issue in external loop of
vrna_subopt*() - SWIG: Add swig class output parameter typemap for Python
- SWIG: Add
__hash__()and__eq__()methods for wrapped_vrna_move_tin Python - SWIG: Return
var_arrayobjects in Python wrappedvrna_neighbors()andvrna_move_neighbor_diff() - SWIG: Refactor file handle wrapping between Python 3 and C
- SWIG: Fix
var_arrayPython slices and associated memory leak - SWIG: Fix bogus
delete/free()calls in swig interface - Add requirements to build
RNAlibwith MSVC for Windows - Remove unused code in
RNApuzzler
- API: Add shift move support to
-
Package
- DOC: Transition reference manual from
doxygentosphinxviabreathebridge - DOC: Merge documentation of C-API and Python API
- DOC: Merge parts of tutorial into reference manual
- AUTOCONF: Refactor autoconf checks for capability to build reference manual
- AUTOCONF: Deactivate build of
RNAxploreriflapackrequirements are not met
- DOC: Transition reference manual from
-
Library
- Make JSON parser integral part of ViennaRNA library
- API: Move modified energy parameters into
modified_baseobject in JSON file(s) - SWIG: Enable stand-alone build of Python interface (for PyPI)
-
Package
- Add enthalpy and terminal end values for predicted stacks with dihydrouridine
- TESTS: Allow for using
pytestto test the Python 3 interface
-
Programs
- Fix preparation of input sequences for modified base support in
RNAcofold
- Fix preparation of input sequences for modified base support in
-
Library
- Fix energy corrections for modified base support when unmodified base is not the same as fallback base, e.g. in the case of inosine
- Add soft constraints to multifold external loop decomposition
- Add soft constraints preparation stage callback
- SWIG: Fix
fc.sc_add_bp()propagation of constraint values - SWIG: Wrap energy parameter file strings
-
Package
- TESTS: Add modified base tests on duplex data with I-C and A-Psi pairs from publications
-
Programs
- Fix double free corruption in
RNAdos - Fix compilation issues due to use of
uintinstead ofunsigned intforRNAxplorer - Fix compilation issues for
RNAxplorerwhen OpenMP is unavailable
- Fix double free corruption in
-
Package
- AUTOCONF: Update autoconf macros
- Update Debian-based packaging rules
This version adds modified base support and a physics-based model to
correct energy contributions depending on the concentration of mono-valent ions
to our implementations. The new features are fully exposed through the command line
tools and the API of RNAlib.
The new implementations for mono-valent ion concentration dependent energy corrections
follows the lines of Einert and Netz, 2011, Theory for RNA folding, stretching, and
melting including loops and salt., Biophys. J. 100, 2745-2753
(https://doi.org/10.1016/j.bpj.2011.04.038)
and is described in Yao et al., 2023, Mono-valent salt corrections for RNA secondary
structures in the ViennaRNA package, currently under revision.
The modified base support that is introduced with this release is based on the soft
constraints framework and at this time integrates nearest neighbor parameters for
the following non-standard nucleotides
- Inosine
- Pseudouridine
- m6A
- 7-deaza-adenosine (7DA)
- Purine (aka nebularine)
- Dihydrouridine
With this release, we started to automatically convert the documentation for the C-API to Python docstrings. As a consequence, we can now provide a Python API documentation at ReadTheDocs.
With this new release, we deactivated building the Python 2 interface
by default. It is still part of the distribution tarball for those that require it
but has to be activated via --with-python2 at configure time.
The list of all major changes in this version can be found below:
-
Programs
- Add modified base input support to
RNAfold,RNAplfold,RNALfold,RNAcofold,RNAsubopt - Fix missing strand separators in
RNAsuboptwhen applied to multiple interacting sequences - Fix sorted output in
RNAsuboptwith--gquadoption - Allow for only
-FpinRNAinverseinstead of always activating-Fm - Fix default value of
RNAinverse -Roption in manpage - Restructure
--*helpoutput and man pages for most executable programs - Allow for mono-valent ion concentration changes in most executable programs (default 1.021M)
- Allow for at least as many threads as CPUs are configured if maximum thread number detection fails
- Fix alignment input parsing in
refold.pl - Add
RNAxplorerprogram to the distribution
- Add modified base input support to
-
Library
- API: Extend
model_detailsto allow for salt concentration changes - API: Add functions for salt concentration change derived energy corrections in
ViennaRNA/params/salt.h - API: Add arbitrary modified base support (
vrna_sc_mod()) via soft constraints mechanism and JSON input data - API: Add Pseuoduridine-A parameters via soft constraints callback
- API: Add Dihydrouridine parameters via soft constraints callback
- API: Add inosine-U and inosine-C parameters via soft constraints callback
- API: Add m6A parameters via soft constraints callback mechanism
- API: Add 7DA modification support via soft constraints
- API: Add Purine (nebularine) modification support
- API: Add new soft constraints multi-callback dispatcher
- API: Add dynamic array data structure utilities
- API: Add string data structure utilities
- API: Add
vrna_strchr()function - API: Fix potential problems in
free_dp_matrices()of LPfold.c - API: Fix z-score initialization in
vrna_Lfoldz()amdvrna_mfe_window_zscore_cb() - API: Fix file close issue in
vrna_file_commands_read() - API: Fix backtracking issue in Zuker subopt
- API: Fix missing soft constraints callback execution in Zuker subopt
- API: Fix enumeration of G-quadruplexes in
vrna_subopt()andvrna_subopt_cb() - API: Fix constraints bug for exterior loop in boltzmann sampling
- API: Allow for enforcing 'must pair' constraint (
|) in dot-bracket constraints strings - API: Fix discrepancy between global and local folding in how hard constraints for unpaired bases and non-specific pairing are applied
- API: Refactor function typdefs to make them actual function pointer typedefs
- SWIG: Fix Python 3 wrapper suffix issue
- SWIG: Fix Perl 5 wrapper for
vrna_ud_prob_get() - SWIG: Only accept upper triangular part of matrix input in
fc.sc_bp_add() - SWIG: Use var_array instead of tuples for Python
RNA.ptable() - SWIG: Add Python wrapper for
vrna_move_neighbor_diff() - SWIG: Add Python docstrings generated from doxygen documentation of C-library
- API: Extend
-
Package
- Update
libsvmto version 3.31 - Update
dlibto version 19.24 - Adapt Debian dependencies
- Fix compilation issues with
RNAforester - AUTOCONF: Fix requirement checks when SVM support is deactivated and swig is missing
- AUTOMAKE: Add
autoparameters for-fltocompile/link flags - AUTOCONF: Require C++17 due to dependencies to compile
DLIB - AUTOCONF: Deactivate Python 2 bindings by default
- Update
Version 2.5.x
-
Programs
- Refactor
ct2dbprogram to allow for pseudoknots in output structure
- Refactor
-
Library
- API: Fix MEA computation for G-quadruplex predictions
- API: Fix memory leak in hard constraints container
- API: Fix RNApuzzler edge-case that resulted in segmentation faults
- API: Fix invalid memory access in
vrna_strjoin() - API: Revisit generic soft constraints for sliding-window base pair probability computations
- API: Enable to overwrite automatic unpaired probability determination in MEA computation
- API: Add
VRNA_PLIST_TYPE_UNPAIREDandVRNA_PLIST_TYPE_TRIPLEidentifiers forvrna_ep_t - API: Add
vrna_init_rand_seed()to initialize RNG with seed - API: Add
vrna_zsc_compute_raw()to obtain mean and sd for Z-score computation - API: Add
vrna_file_connect_read_record()function to parse connectivity table (*.ct) files - API: Add
vrna_strtrim()function - API: Update sanity checks for input in
vrna_pbacktrack_sub*() - API: Allow for pseudo-knots in
vrna_db_from_ptable() - API: Do not use
min_loop_size = 0for multi strand interaction prediction - API: Remove unnecessary uses of
min_loop_sizeat multiple locations - API: Deprecate cutpoint member of vrna_fold_compound_t and prepare for 5'/3' encoding
- API: Refactor sequence addition/preparation for
vrna_fold_compound_t - DOC: Update documentation
- SWIG: Add simple dot-plot file wrapper
plot_dp_EPS() - SWIG: Add
sequence,sequence_encodingandsequence_encoding2attributes tofold_compoundobjects - SWIG: Fix RNG wrapping and initialize RNG upon module load and update associated functions
- SWIG: Add more access to member variable arrays for various objects used throughout the library
- SWIG: Add memory efficient wrapper for dynamically allocated arrays and matrices
- SWIG: Shadow pair table data structure for efficient interactions between C and target languages
- SWIG: Expose hard constraints members in
fold_compoundobjects - SWIG: Add
exp_E_ext_stem()method (vrna_exp_E_ext_stem()) tofold_compoundobjects - SWIG: Expose DP matrices within
fold_compoundobjects - SWIG: Fix memory leak in wrapper for
vrna_db_from_ptable()
-
Package
- Update dlib to version 19.23
- DOC: Update doxygen.conf for version 1.9.2
- AUTOCONF: Factor-out Naview layout algorithm to allow for deactivating the Naview layout algorithm at configure-time
- AUTOCONF: Make LaTeX checks more portable and update LaTeX package checks
- AUTOCONF: Check whether we can build the swig interface when SVM support is deactivated
- AUTOCONF: Fix condition check for CLA build
This version adds multi-strand RNA interaction support to our implementations,
exposed to end-users in the form of the novel command line tool
RNAmultifold and corresponding changes in the API of RNAlib.
The new implementations for RNA-RNA interaction can now handle multiple RNA input
strands that shall form interaction complexes. For that, we implemented a generalization
of the concatenation approach of RNAcofold following the ideas of
Dirks et al. 2007, Thermodynamic Analysis of Interacting Nucleic Acid Strands,
SIAM REVIEW Vol. 49, No. 1, pp. 65-88
https://doi.org/10.1137/060651100 and
Lorenz et al., 2021, Efficient Algorithms for Co-Folding of Multiple RNAs, In:
Ye X. et al. (eds) Biomedical Engineering Systems and Technologies. BIOSTEC 2020.
Communications in Computer and Information Science, vol 1400. Springer, Cham.
https://doi.org/10.1007/978-3-030-72379-8_10.
The major new features for multiple interacting RNAs are
- MFE prediction
- Partition function and base pair probability computation
- Equilibrium concentration computation
- Suboptimal structure enumeration within an energy band around the MFE
- Suboptimal structure enumeration ala Zuker 1989
- Free energy evaluation
-
Due to the new implementations, the output order for ensemble free energies of
interacting complexes slightly differ between
RNAcofoldandRNAmultifoldwith two input sequences. Apart from that,RNAmultifoldcan be considered a drop-in replacement forRNAcofold.
With this new release, we also adapted the covariance score computation for the prediction
of consensus structures of multiple sequence alignments. In particular, the condition that
decides which columns of the alignment are allowed to pair was updated to yield more sane
results for edge-cases with many gaps and non-canonical base pairs (counter-examples). Therefore,
comparing the output of RNAalifold against that of version 2.4.18 or less will
most likely result in differences.
This version also makes Python 3 the default Python for the SWIG interface.
The only difference you should see, however, is that the source tree changed from directories
interfaces/Python and interfaces/Python3 to interfaces/Python and
interfaces/Python2.
Additionally, we add PDF versions of our
contributors license agreement (CLA) to the source
code tar-ball.
The list of all major changes in this version can be found below:
-
Programs
- Add
RNAmultifoldprogram to compute secondary structures for multiple interacting RNAs - Add multistrand capabilities to
RNAeval - Add multistrand capabilities to
RNAsubopt - Replace
RNAcofoldwith a wrapper toRNAmultifold - Fix computation of BB homodimer base pair probabilities in
RNAcofold
- Add
-
Library
- API: Fix use of undefined values in deprecated function
PS_dot_plot() - API: Fix probability computations for unstructured domains within multibranch loops
- API: Fix index error in ensemble defect computations
- API: Fix hard constraints behavior on non-specific base pairing
- API: Fix segmentation fault for short input sequences in
vrna_hx_from_ptable() - API: Fix memory leak in static
rna_layout()function - API: Fix corner-case in covariance score computation on sequence alignments that determines which alignment columns may pair and which don't
- API: Add MFE computations for multiple interacting strands
- API: Add partition function computations for multiple interacting strands
- API: Add base pair probability computations for multiple interacting strands
- API: Add suboptimal structure prediction for multiple interacting strands
- API: Add multistrand capabilities to
vrna_eval*()functions - API: Add new function
vrna_equilibrium_conc()for concentration dependency computations of multiple interacting strands withdlibbackend - API: Add
vrna_equilibrium_constants()function to obtain equilibrium constants for different complexes of multiple interacting strands - API: Add function
vrna_pf_add()to add ensemble free energies of two ensembles - API: Add function
vrna_pf_substrands()to get ensemble free energies for complexes up to a specific number of interacting strands - API: Add function
vrna_n_multichoose_k()to obtain a list of k-combinations with repetition - API: Add
vrna_cstr_discard()function to allow for discarding char streams prior to flushing - API: Add
vrna_bp_distance_pt()function to allow for base pair distance computation with pseudo-knots - API: Add functions
vrna_pbacktrack_sub*()to allow for stochastic backtracing within arbitrary sequence intervals - API: Add functions
vrna_boustrophedon()andvrna_boustrophedon_pos()to generate lists of or obtain values from sequences of Boustrophedon distributed integer numbers - API: Add
vrna_pscore()andvrna_pscore_freq()functions to obtain covariance score for particular alignment columns - API: Rewrite Zuker suboptimals implementation
- API: Remove old cofold implementations
- API: Make
typeattribute ofvrna_mx_mfe_tandvrna_mx_pf_ta constant - API: Guard more functions in
utils/structure_utils.cagainstNULLinput - API: Rename
vrna_E_ext_loop()tovrna_eval_ext_stem() - API: Use v3 typedefs in dot-plot function declarations
- SWIG: Fix Python 3 file handle as optional argument in
eval*functions and methods - SWIG: Add wrapper for
vrna_pf_add() - SWIG: Add wrapper for
vrna_hx_from_ptable() - SWIG: Add wrapper for
vrna_db_from_probs()
- API: Fix use of undefined values in deprecated function
-
Package
- Update
libsvmto version 3.25 - Make Python 3.x the default Python for the scripting languange interfaces
- Add Python3 capability for Mac OS X installer builds
- TESTS: Create TAP driver output for all unit tests (library, executables, SWIG interfaces)
- Remove compile-time switch to deactivate Boustrophedon backtracing scheme (this is the status-quo now)
- Add Contributors License Agreement (CLA) to the Package in
doc/CLA/
- Update
Version 2.4.x
-
Programs
- Fix and refactor
RNApkplexprogram - Fix occasional backtracing errors in
RNALalifold - Restrict available dangling end models in
RNALalifoldto 0 and 2 - Prevent segmentation faults upon bogus input data in
RNAfold,RNAalifold,RNAcofold,RNAheat, andRNAeval - Free MFE DP matrices in
RNAsuboptBoltzmann sampling when not required anymore
- Fix and refactor
-
Library
- API: Add
vrna_abstract_shapes()andvrna_abstract_shapes_pt()functions to convert secondary structures into their respective abstract shape notation ala Giegerich et al. 2004 - API: Add functions
vrna_seq_reverse()andvrna_DNA_complement()to create reverse complements of a sequence - API: Add more soft constraint handling to comparative structure prediction
- API: Add generic soft constraints for sliding window comparative MFE backtracing
- API: Add
vrna_ensemble_defect_pt()that accepts pair table input instead of dot-bracket string to allow for non-nested reference structures - API: Add failure/success return values to generic soft constraints application functions
- API: Refactor
RNAPKpleximplementation by better using constraints framework and moving out many parts fromRNAPKplex.cintoRNAlibas separate re-usable functions - API: Fix energy contributions used in
RNAPKpleximplementations - API: Fix energy evaluation for cofolding with dangle model 1
- API: Fix wrong arithmetic usage for PF variant of combined generic and simple soft constraints applied to external loops
- API: Fix memory size in #vrna_fold_compound_t initialization
- API: Fix bogus memory access for comparative prediction when preparing hard constraints
- API: Fix wrong index usage in hard constraints for comparative base pair probability computations of internal loops
- API: Fix G-Quadruplex contributions as part of multibranch loops in single sequence base pair probability computations
- API: Fix multibranch loop MFE decomposition step for multiple strand cases
- API: Fix external loop generic hard constraint index updating for partition function computations
- API: Fix memory allocation for auxiliary grammar data structure
- API: Fix incorporation of auxiliary grammar contrib for closing pairs in sliding-window MFE computation
- API: Fix DP matrix intitialization in sliding window MFE computations (fixes occasional backtracing issues in comparative sliding-window MFE computations)
- API: Make
vrna_sc_t.typeattribute a constant - API: Remove upper-triangular hard constraint matrix in favor of full matrix
- API: Always ensure sane base pair span settings after
vrna_fold_compound_prepare() - API: Return INF on predictions of
vrna_mfe_dimer()that fail due to unsatisfiable constraints - API: Rename internally used hard and soft constraints API symbols
- API: Fix header file inclusions to prevent #include cycles
- SWIG: Add wrapper for
vrna_file_fasta_read_record() - SWIG: Fix memory leak in wrapper for
vrna_probs_window() - SWIG: Refactor and therefore fix soft constraint binding functions for use in comparative structure predictions
- SWIG: Fix typo that prevented properly wrapping
vrna_params_load_RNA_Andronescu2007() - SWIG: Unify wrappers for
vrna_ptable()andvrna_ptable_from_string()
- API: Add
-
Package
- REFMAN: Refactored structure annotation documentation
- REFMAN: Update Mac OS X install section
- Replace
DEFplaceholders in energy parameter files with their value of -50 - Update
RNAlocminsubpackage to properly compile with more stringent C++ compilers - Update
RNAforestersubpackage to properly compile with more stringent C++ compilers - Update autotools framework, e.g. checks for pthreads
- Update universal binary build instructions for Mac OS X builds to enable ARM compilation for M1 CPUs
- Source code changes since v2.4.17 are available on Github.
-
Programs
- Fix
RNAup -bmode with shorter sequence first - Add
--backtrack-globaloption toRNALfold(currently only available for dangles == 2 | 0) - Add
--zscore-pre-filterand--zscore-report-subsumedoptions toRNALfold
- Fix
-
Library
- API: Fix multiloop backtracing with soft constraints for unpaired positions in
vrna_subopt()andvrna_subopt_cb() - API: Fix parameter parse in
vrna_params_load_from_string() - API: Add
vrna_heat_capacity()andvrna_head_capacity_cb()functions to RNAlib - API: Add backtracing function
vrna_backtrack_window()for global MFE structure to sliding-window predictions - API: Add SVG support for
RNApuzzlerstructure layouts - API: Make
vrna_md_targument to vrna_fold_compound() a constant pointer - API: Remove missing symbols from header file
ViennaRNA/params/default.h - API: Refactor z-score threshold filter handling for sliding-window MFE prediction
- SWIG: Fix typo in interface functions to load DNA parameters
- SWIG: Add python-3.9 autoconf checks
- SWIG: Add
vrna_head_capacity*()wrappers - SWIG: Add access to raw energy parameters
- SWIG: Add
aliasandpairattribute to objects of typemd - SWIG: Add out/varout typemaps for 2-dimensional int-like arrays
- SWIG: Add all data fields to objects of type
paramandexp_param
- API: Fix multiloop backtracing with soft constraints for unpaired positions in
-
Package
- Fix Debian and Windows installer files
-
Programs
- Fix backtracing errors in
RNALalifoldfor alignments with more than32768columns - Fix backtracing errors in
RNAalifoldandRNALalifoldfor rare cases when two alignment columns may pair due to covariance score threshold but still yield infinite energies due to energy model - Refactored manpages/help options for
RNAplfold,RNAplot,RNApvmin,RNAsubopt, andRNAup
- Fix backtracing errors in
-
Library
- API: Fix undefined behavior due to short int overflows when accessing alignment lengths with alignments larger than 32768 columns. This fixes occasional backtracing errors in
RNALalifoldandvrna_mfe_window() - API: Fix adding pscore to base pairs that yield INF energy in comparative global and local MFE prediction
- API: Add
vrna_convert_kcal_to_dcal()and vice-versa function for safely converting integer to float representations of energy values - SWIG: Add a reasonable
Pythoninterface for objects of typevrna_path_t - SWIG: Add a wrapper for
vrna_seq_encode()
- API: Fix undefined behavior due to short int overflows when accessing alignment lengths with alignments larger than 32768 columns. This fixes occasional backtracing errors in
-
Package
- Move
units.hinclude file toViennaRNA/utils/units.h
- Move
-
Programs
- Fix compilation of
Kinfoldwith GCC 10 - Add
--en-onlyflag toRNAsuboptto allow for sorting by energy only - Prevent
RNAcofoldto process input with more than two strands - Add cutpoint marker to dot-plots created with
RNAcofold -a - Update
Kinfoldto version1.4
- Fix compilation of
-
Library
- API: Fix removal of strand delimiter in
vrna_plot_dp_PS_list() - API: Fix
vrna_enumerate_necklaces() - API: Fix bogus backtracing for co-folded structures in
vrna_subopt()andvrna_subopt_cb() - API: Fix storing co-folded structures for sorted output in
vrna_subopt() - API: Fix multibranch loop component hard constraints for multi-strand cases
- API: Prevent adding internal loop energy contributions to enclosed parts with
energy=INF - API: Adapt
vrna_db_pack()/vrna_db_unpack()functions to produce comparable strings - API: Add sorting modes
VRNA_UNSORTED,VRNA_SORT_BY_ENERGY_LEXICOGRAPHIC_ASC, andVRNA_SORT_BY_ENERGY_ASCtovrna_subopt() - API: Add
vrna_strjoin()function - API: Add missing case to external loop hard constraints
- API: Make hard constrains strand-aware
- SWIG: Fix invalid memory access when using
MEA_from_plist()inPerl 5orPython - SWIG: Enable keyword argument features in
Pythoninterface of constructors forfold_compound,md,move,param, andexp_paramobjects - SWIG: Enable autodoc feature for
Pythoninterface of constructors forfold_compound,md, andmoveobjects - SWIG: Enable
toStringconversion forPythoninterface for objects of typefold_compound,md,move,params,exp_params, andsubopt_solution - SWIG: Add (read-only) attributes
type,length,strands,params, andexp_paramsto objects of typefold_compound - SWIG: Make attributes of objects of type
paramandexp_paramread-only - Add array of strand nicks to EPS dot plot files instead of single cutpoint
- Draw separator line for each strand nick in EPS dot-plots
- Update
libsvmto version 3.24
- API: Fix removal of strand delimiter in
-
Package
- Disable Link-Time-Optimization (
LTO) for third-party programs linking againstRNAlibusingpkg-config - TESTS: Fix results dir path for out-of-tree builds
- TESTS: Set default timeout for library tests to 20s
- Disable Link-Time-Optimization (
-
Programs
- Fix
RNApvminpertubation vector computation - Add non-redundant sampling option to
RNApvmin - Add
RNAdosprogram to compute density of states - Add
-P DNAconvenience command line parameter to most programs to quickly load DNA parameters without any input file - MAN: Add example section to man-page of
RNAalifold
- Fix
-
Library
- API: Fix memory leak in
vrna_path_gradient() - API: Fix release of memory for
vrna_sequence_remove_all() - API: Fix soft-constraints application in
vrna_sc_minimize_pertubation()that prevented proper computation of the pertubation vector - API: Add 5' and 3' neighbor nucleotide encoding arrays and name string to
vrna_seq_t - API: Add new data structure for multiple sequence alignments
- API: Add
vrna_sequence_order_update()function - API: Add non-redundant sampling mode to
vrna_sc_minimize_pertubation()through passing negative sample-sizes - API: Add v3.0 API functions for maximum expected accuracy (MEA) computation
- API: Include energy parameter sets into
RNAliband provide functions to load them at runtime - API: Prepare sequence data in
vrna_fold_compound_twithvrna_sequence_add() - API: Use
vrna_pbacktrack_num()instead ofvrna_pbacktrack()invrna_sc_minimize_pertubation()to speed-up sample generation - Reduce use of global variable
cut_pointinRNAlib - SWIG: Use
importlibin favor ofimpto determine Python 3 tag extension - SWIG: Update various wrapper functions
- SWIG: Add wrappers for MEA computation with
vrna_MEA()andvrna_MEA_from_plist - SWIG: Add wrappers for
vrna_pr_structure()andvrna_pr_energy()
- API: Fix memory leak in
-
Package
- REFMAN: Fix LaTeX code in
units.hthat prevented proper compilation withpdflatex - Add an R script to create 2D landscape plots from
RNA2Dfoldoutput - Add
gengetoptto configure-time requirements to build man-pages - Add new energy parameter file
rna_misc_special_hairpins.parwith additional UV-melting derived parameters for Tri- and Tetra-loops - Update RNA Tutorial
- Colorize final configure script message
- REFMAN: Always use
pdflatexto compile reference manual and tutorial - EXAMPLES: Add Python script that performs computations equivalent to
RNAfold -p --MEA
- REFMAN: Fix LaTeX code in
-
Programs
- Fix centroid structure prediction for
RNAcofold - Fix
--noLPoption forRNALalifold
- Fix centroid structure prediction for
-
Library
- API: Refactor and fix collision handling in
vrna_hash_table_t - API: Fix one access using wrong index for odd dangles in
loops/external.c - API: Add two missing
MLbasecontributions for MFE prediction inloops/multibranch.c - API: Refactor multiloop MFE backtracking for odd dangles
- API: Add function
vrna_backtrack5()to allow for MFE backtracking of sub-sequences starting at the 5'-end - API: Reduce usage of global macro
TURNby replacing it withmin_loop_sizefield ofvrna_md_t - API: Add functions
vrna_path_direct()andvrna_path_direct_ub()that may also return move lists instead of dot-bracket lists - API: Add functions
vrna_pt_pk_remove()andvrna_db_pk_remove()that remove pseudoknots from an input structure - API: Fix invalid memory access for lonely pair mode (
--noLP) in comparative sliding-window MFE prediction - SWIG: Fix access to global variable
pf_smoothandpf_smoothattribute inmodel_detailsobject - SWIG: Fix Python reference counting for
Py_Noneininterfaces/findpath.iwrapper - SWIG: Refactor reference counting for all Python2 and Python3 wrappers
- REFMAN: Larger updates and restructuring of reference manual
- API: Refactor and fix collision handling in
-
Package
- Install example scripts and source code files, e.g. to $prefix/share/ViennaRNA/examples
- Properly pass GSL, PTHREADS, and MPFR flags to sub-projects
- Fix
RNApuzzlerheader file installation - SWIG: Include Python 3.7 and 3.8 in list of autoconf-probed python interpreters
- SWIG: Fix wrapper building for
swig >= 4.0.0
-
Programs
- Add non-redundant stochastic backtracing option for
RNAalifold - Add
--noDPoption to suppress dot-plot output inRNAfoldandRNAalifold - Add
RNApuzzler(4) andRNAturtle(3) secondary structure layout algorithm options toRNAfoldandRNAplot - Update help/man page of
RNALfold - Allow for multiple input files and parallel input processing in
RNAheat
- Add non-redundant stochastic backtracing option for
-
Library
- API: Fix declaration of
vrna_move_apply_db() - API: Fix
vrna_path()lexicographical ordering in gradient walks - API: Enable non-redundant stochastic backtracing for comparative structure prediction
- API: Enable stochastic backtracing for circular comparative structure prediction
- API: Enable stochastic backtracing of subsequences (5' prefixes) for comparative structure prediction
- API: Add
pf_smoothattribute tovrna_md_tdata stucture to allow for disabling Boltzmann factor energy smoothing - API: Add functions to allow for resuming non-redundant stochastic backtracing
- API: Add functions to retrieve multiple stochastically backtraced structures (list and callback variants)
- API: Add
vrna_positional_entropyto compute vector of positional entropies - API: Add
RNApuzzlerandRNAturtlesecondary structure layout algorithm (Wiegreffe et al. 2018) - API: Add v3.0 API for secondary structure layout/coordinate algorithms
- API: Add more helper/utility functions for
vrna_move_tdata structures - API: Add callback-based neighborhood update function for (subsequent)
vrna_move_tapplication - API: Add abstract heap data structure available as
<ViennaRNA/datastructures/heap.h> - API: Refactor and speed-up gradient walk implementation available as
vrna_path_gradient() - API: Substitute
vrna_file_PS_aln_sub()alignment plot function byvrna_file_PS_aln_slice()that actually slices out a sub-alignment - API: Rename
vrna_annotate_covar_struct()tovrna_annotate_covar_db()and add new functionvrna_annotate_covar_db_extended()to support more bracket types - API: Calling
vrna_params_reset()now implies a call tovrna_exp_params_reset()as well - API: Move landscape implementations into separate directory, thus headers should be included as
<ViennaRNA/landscape/move.h>,<ViennaRNA/landscape/neighbor.h>, etc. - Ensure proper rescaling of energy parameters upon temperature changes
- Refactor soft constraints implementation in stochastic backtracing
- SWIG: Wrap all non-redundant stochastic backtracing functions to scripting language interface(s)
- SWIG: Refactor stochastic backtracing interface(s)
- SWIG: Add proper constructor for objects of type
vrna_ep_t - SWIG: Sanitize alignment plot function interface(s)
- API: Fix declaration of
-
Package
- Update Ubuntu/Debian and OpenSUSE build instructions
- Reduce intra-package dependency on non-v3.0 API
-
Programs
- Add
--commandsoption toRNAsubopt - Add non-redundant Boltzmann sampling mode for
RNAsubopt
- Add
-
Library
- API: Fix wrong access to base pair soft constraints in equilibrium probability computations
- API: Fix behavior of
vrna_nucleotide_encode()with lowercase characters in sequence - API: Fix behavior of
encode_char()with lowercase characters in sequence - API: Fix forbidden
GUpairs behavior inpscorecomputation for comparative folding - API: Fix potential errors due to uninitialized
nextpointers invrna_move_tofvrna_eval_move_shift_pt - API: Add
AVX 512optimized version of MFE multibranch loop decomposition - API: Add functions for CPU SIMD feature detection
- API: Add dispatcher to automatically delegate exterior-/multibranch loop MFE decomposition to supported SIMD optimized implementation
- API: Add function
vrna_dist_mountain()to compute mountain distance between two structures - API: Add function
vrna_ensemble_defect()to compute ensemble defect given a target structure - API: Add non-redundant Boltzmann sampling
- API: Change behavior of
vrna_cstr_free()andvrna_cstr_close()to always flush output before unregistering the stream - SWIG: Add interface for
vrna_loopidx_from_ptable()
-
Package
- Activate compilation for compile-time supported SIMD optimized implementations by default
- Replace
--enable-sseconfigure script option with--disable-simd
-
Programs
- Fix wrong output filename for binary opening energies in
RNAplfold - Enable G-Quadruplex support for partition function computation in
RNAalifold
- Fix wrong output filename for binary opening energies in
-
Library
- Fix broken
SSE4.1support for multibranch loop MFE computation that resulted in increased run times - Fix redundant output issue in subopt backtracking with unusually high delta energies (≥INF)
- Restore default behavior of
'|'symbol in dot-bracket hard constraint strings that got lost with version 2.2.0 - Add faster (cache-optimized) version of Nussinov Maximum Matching algorithm
- Change default linker- and loop length computations for G-Quadruplex predictions in comparative prediction modes
- Add hard constraints warning for base pairs that violate the
min_loop_sizeof the model - Update
libsvmto version 3.23 - API: Add functions to set auxiliary grammar extension rules
- API: Replace upper-triangular hard constraints matrix with full matrix for cache-optimized access
- API: Add G-Quadruplex prediction support for comparative partition function
- API: Remove
VRNA_GQUAD_MISMATCH_PENALTYandVRNA_GQUAD_MISMATCH_NUM_ALImacros - SWIG: Fix invalid memory access in
subopt()method of fold_compound object when writing to file - SWIG: Add wrapper for Nussinov Maximum Matching algorithm
- Fix broken
-
Package
- Add
-ftree-vectorizecompile flag by default if supported
- Add
-
Programs
- Fix interactive mode behavior for multiple sequence alignment input in
RNAalifold,RNALalifold - Allow for Stockholm formatted multiple sequence alignment input in
RNAevalandRNAplot - Allow for multiple input files in
RNAevalandRNAplot - Allow for parallel processing of input batch jobs in
RNAevalandRNAplot - Add
-g / --gquadoption to activate G-Quadruplex support inRNAheat - Warn on unsatisfiable hard constraints from dot-bracket string input in
RNAfold,RNAcofold, andRNAalifold
- Fix interactive mode behavior for multiple sequence alignment input in
-
Library
- Fix parameter order bug in
vrna_path_findpath*functions that resulted in too large search widths - Fix wrong application of base pair soft constraints in partition function computations
- Fix position ruler string in EPS alignment output files
- Fix MFE backtracking errors that might appear under specific hard constrained base pair patterns
- Complete soft constraints additions to Boltzmann sampling implementation for single sequences
- API: Refrain from reading anything other than
#=GC SS_consto retrieve structures when parsing Stockholm 1.0 format - API: Allow for disabling alignment wrapping in
vrna_file_PS_aln*functions - API: Do not remove G-Quadruplex annotation from WUSS formatted structure strings upon calls to
vrna_db_from_WUSS() - API: Enable G-Quadruplex related average loop energy correction terms in verbose output of
vrna_eval_*functions - API: Speed-up backward compatibility layer for energy evaluation functions that unnecessarily slowed down third-party tools using the old API
- API: Allow for passing dot-bracket strings with
&strand-end identifier to simplevrna_eval_*functions - API: Remove implicit
exit()calls from global MFE backtracking implementation.
- Fix parameter order bug in
-
Programs
- Fix compilation of
RNAforesterwith C++17 standard - Fix tty input detection in
RNAcofold - Fix bad memory access with
RNAcofold -p
- Fix compilation of
-
Library
- API: Fix incorrect unpaired probability computations in
vrna_probs_window() - API: Fix potential out-of-bounds access situations (for circular RNA folding) in
eval.c - API: Fix comparative exterior internal loop partition function computation for circfold
- SWIG: Fix false-positive use of uninitialized value in
Python3/file_py3.i
- API: Fix incorrect unpaired probability computations in
-
Package
- TESTS: Add tests for special features in
RNAalifold - TESTS: Add test case for
RNAcofold -p
- TESTS: Add tests for special features in
-
Programs
- Allow for parallel processing across multiple input files in
RNAfold - Allow for arbitrary number of input files in
RNAalifold - Allow for parallel processing of input data in
RNAalifold - Allow for arbitrary number of input files in
RNAcofold - Allow for parallel processing of input data in
RNAcofold - Enable parallel processing in
RNAfold,RNAcofold,RNAalifoldfor MS Windows build - Add centroid and MEA structure computation to
RNAcofold - Include ligand binding energies in centroid and MEA structure output of
RNAfold - Refactor
ct2dbprogram to process multiple structures from single.ctfile
- Allow for parallel processing across multiple input files in
-
Library
- API: Enable processing of comparative fold_compound with
vrna_pr_*()functions - API: Refactor
vrna_ostream_tto enableNULLinput invrna_ostream_provide() - API: Major refactoring in loop energy evaluations (MFE and PF)
- API: Make
vrna_mx_pf_aux_el_tandvrna_mx_pf_aux_ml_sopaque pointers - API: Make fold_compound field
typeaconstattribute - API: Refactor MFE post-processing for circular RNAs
- API: Add motif name/id support for unstructured domains
- API: Remove major part of implicit
exit()calls in RNAlib - API: Add implementations of Boyer-Moore-Horspool search algorithm
- API: Add implementations to determine number of rotational symmetry for strings (of objects)
- API: Make
vrna_cmd_tan opaque pointer - API: Move headers for constraints, datastructures, io, loop energy evaluation, energy parameters, plotting, search, and utilities into separate subdirectories (backward compatibility is maintained)
- API: Add hash table data structure
- API: Fix discrepancy between comparative and single sequence
--noLPpredictions - API: Add functions to replace 'old API' interface of
RNAstruct.h - API: Add functions to replace 'old API' interface of
aln_util.h - API: Add generic soft constraints support to suboptimal structure prediction sensu Wuchty et al.
- SWIG: Refactor callback execution for Python 2 / 3 interface to reduce overhead
- SWIG: Fix configure-time check for Python 3 interface build
- SWIG: Fix Python 3 IO file stream to C
FILE *conversion
- API: Enable processing of comparative fold_compound with
-
Package
- Add configure time check for LTO capabilities of the linker
- Cosmetic changes in final configure notice
- Major changes in source tree structure of the library
- Add autoconf checks for maintainer tools
- Generate C strings from static PostScript files at configure time (for structure- and dot plots)
- REFMAN: Large updates in API documentation and structure of reference manual
This version fixes two regressions in comparative structure prediction.
In particular, we adapt the computation of base pairing probabilities and Boltzmann sampling to the changes introduced with Version 2.4.4, and restore dot-bracket based hard constrained base pairs that got lost with Version 2.4.2.
-
Programs
- Stabilize rounding of free energy output in
RNAalifold
- Stabilize rounding of free energy output in
-
Library
- API: Fix potential rounding errors for comparative free energies in
eval.candmfe.c - API: Fix regression in exterior loop dangling end contributions for comparative base pair probabilities and Boltzmann sampling
- API: Fix regression with hard constrained base pairs for comparative structure prediction
- API: Fix potential rounding errors for comparative free energies in
-
Package
- TESTS: Add basic tests for
RNAalifoldexecutable - TESTS: Ignore 'frequency of MFE structure' in
RNAcofoldpartition function checks
- TESTS: Add basic tests for
-
Programs
- Allow for arbitrary number of input files in
RNAfold - Allow for parallel processing of input data in
RNAfold(unavailble for Windows platform) - Add SHAPE reactivity support through commandline options for
RNAplfold - Fix unstructured domain motif detection in MFE, centroid, and MEA structures computed by
RNAfold - Limit allowed set of commands in command file for
RNAcofoldto hard and soft constraints
- Allow for arbitrary number of input files in
-
Library
- API: Add functions to compute equilibrium probability of particular secondary structures
- API: Add dynamic string stream data type and associated functions
- API: Add priority-queue like data structure with unordered fill capability and ordered output callback execution
- API: Add functions to detect unstructured domain motifs in MFE, centroid, and MEA structures
- API: Fix bug in sliding-window partition function computation with SHAPE reactivity and Deigan et al. conversion method
- API: Fix application of
'<'and'>'constraint symbols in dot-bracket provided constraints - API: Fix MEA structure computation in the presence of unstructured domains
- API: Stabilize order of probability entries in EPS dot-plot files
- Fix compiler warnings on wrong type of
printf()innaview.c - Define
VRNA_VERSIONmacro as string literal and add macros for major, minor, and patch numbers - SWIG: Fix 'next' is a perl keyword warnings for Perl5 wrapper
- SWIG: Catch errors and throw execptions whenever scripting language provided callback functions are not applicable or fail
- SWIG: Add keyword arguments and autodoc feature for Python/Python3 wrappers
-
Package
- Stabilize parallel make of Mac OS X installer
- Add energy parameter set from Langdon et al. 2018
- Add autoconf checks for POSIX threads compiler/linker support
This version introduces a correction with respect to the handling of dangling end contributions in exterior
loops when computing the partition function for multiple sequence alignments!
Therefore, the ensemble free energies
and equilibrium probabilities computed with programs such as RNAalifold and RNAplfold
may slightly differ compared to previous versions.
The above correction can briefly be described as a harmonization of the partition function model compared to the MFE implementation, where end-gaps in the alignment can never contribute any stability to helices branching off the exterior loop.
-
Programs
- Change verbose output for soft-constraints derived ligand binding motifs in
RNAfold - Allow for lowercase letters in
ct2dbinput - Fix annotation of PostScript output for soft-constraint derived ligand binding motifs in
RNAfold - Fix sporadic segmentation faults for
RNAsuboptin cofold mode
- Change verbose output for soft-constraints derived ligand binding motifs in
-
Library
- Add simplified interface for
vrna_pf_dimer() - Add new API functions for exterior loop evaluations
- Add simplified interfaces for energy evaluation with G-Quadruplexes and circular RNAs
- Add
vrna_path_findpath()functions that allow for specification of an upper bound for the saddle point - Add automatic CPP suggestions for deprecated function substitutes
- Fix dangling end contributions in comparative partition function for exterior loops
- Fix bug in interior-like G-Quadruplex MFE computation for single sequences
- Fix bug in
eval_int_loop()that prevented propagation of energy evaluation for loops with nick in strands - Fix several bugs for SHAPE reactivity related comparative partition function computations
- Fix constraint indices for multibranch loops in unpaired probability computations of
LPfold.c - Move concentraton dependent implementation for co-folding to separate compile unit
- Major restucturing and constraints feature additions in loop type dependent energy evaluation functions
- Major restructuring in MFE implementations
- Major restructuring in PF implementations
- Minor fixes in Boltzmann sampling implementation
- SWIG: Fix wrappers for
vrna_path_findpath()implementation - SWIG: Add tons of energy evaluation wrappers
- SWIG: Fix configure-time check of Perl5 interface build capabilities
- SWIG: Wrap functions from
walk.candneighbor.c - SWIG: Add configure-time linker check for Python3 interface
- Add simplified interface for
-
Package
- Add some missing references to manpages of executable programs
- Heavy re-ordering of the
RNAlibreference manual - Fix autoconf switch to enable deprecation warnings
-
Programs
- Add
--pf_scalecommandling parameter toRNAplfold - Prevent
RNAplfoldfrom creatinginf/-infoutput when solution set is empty with particular hard constraints - Include
RNAforesterv2.0.1 (minor changes in code base to make modern compilers happier) - Fix probability computation for stochastic backtracking in
RNAsubopt --stochBT_enoutput
- Add
-
Library
- Add constraint framework for single sequence circular RNA structure prediction
- Stabilize partition function scaling by always using sfact scaling factor from model details
- Fix handling of dangling end contribution at sequence boundaries for sliding window base pair probability computations
- Fix handling of base pair hard constraints in sliding-window implementations
- Fix sliding-window pair probability computations with multibranch-loop unpaired constraints
- Fix sliding-window non-specific base pair hard constraint implementation
- Fix regression in comparative structure prediction for circular RNAs
-
Package
- Add
RNAfoldtest suite to check for working implementation of constraints for circular RNAs - Add a brief contribution guideline
CONTRIBUTING.md - Fix
LDFLAGSfor scripting language interfaces in corresponding Makefiles
- Add
-
Programs
- Fix regression in
RNAupunstructuredness and interaction energy computations - Fix build problems of
RNAlocminwith older compilers - Fix bad memory access in
RNAsuboptwith dot-bracket constraint - Add full WUSS support for
--SS_consconstraint option inRNAalifold - Add commandline option to
RNALalifoldthat enables splitting of energy contributions into separate parts - Add CSV output option to
RNAcofold - Use the same model details for SCI computations in
RNAalifold - Use original MSA in all output generated by
RNAalifoldandRNALalifold
- Fix regression in
-
Library
- Fix G-Quadruplex energy corrections in comparative structure energy evaluations
- Fix discrepancy in comparative exterior loop dangling end contribution of eval vs. MFE predictions
- Fix sequence length confusions when FASTA input contains carriage returns
- Fix sliding-window hard constraints where single nucleotides are prohibited from pairing
- Fix dot-bracket output string length in sliding-window MFE with G-Quadruplexes
- Fix unpaired probability computations for separate individual loop types in
LPfold.c - Add missing hard constraint cases to sliding-window partition function implementation
- Abort computations in
vrna_eval_structure_v()if structure has unexpected length - API: Add new functions to convert dot-bracket like structure annotations
- API: Add various new utility functions for alignment handling and comparative structure predictions
- API: Add function
vrna_strsplit()to split string into tokens - API: Do not convert sequences of input MSA to uppercase letters in
vrna_file_msa_read_record() - API: Rename
vrna_annotate_bp_covar()andvrna_annotate_pr_covar() - API: Add new noLP neighbor generation
- SWIG: Add wrapper for functions in
file_utils_msa.h - SWIG: Add wrappers for
vrna_pbacktrack()andvrna_pbacktrack5() - SWIG: Add
vrna_db_to_element_string()to scripting language interface
-
Package
- REFMAN: Fix formula to image conversion in HTML output
-
Programs
- Regression: Fix homo-dimer partition function computation in
RNAcofold - Add SHAPE reactivity support to
RNAcofold - Add SHAPE reactivity support to
RNALalifold
- Regression: Fix homo-dimer partition function computation in
-
Library
- Regression: Fix reverting
pf_scaleto defaults aftervrna_exp_params_rescale() - Fix memory leak in SWIG generated
fold_compoundmethods returning dot-bracket strings - Fix memory leaks in
double **returning functions of SWIG Perl5 interface - Fix memory leak in
vrna_ep_t.__str__()function of SWIG interface
- Regression: Fix reverting
-
Package
- Add unit tests for
RNAcofoldexecutable
- Add unit tests for
This version adds hard/soft constraints support for our implementations of
sliding-window structure prediction as used in the executable programs
RNALfold and RNAplfold.
Together with the new feature of hard- and soft-constraints for sliding-window
structure prediction we unified and corrected the implementations used in the
tools RNALfold, RNALalifold, and RNAplfold.
The major changes are
-
All the above tools now use the same definition of maximum base pair
span.
This means that for a maximum base pair span of L the recursions only consider base pairs (i, j) with (j - i + 1) ≤ L. -
Locally optimal structures that are part of another, larger locally
optimal structure are filtered out.
This mainly affects the output ofRNALfoldthat sometimes did not filter properly. Note, however, that filtering is only performed for consecutive hits!. Thus, it may still happen that locally optimal substructures appear in the output. -
Locally optimal structure must not be branched in the exterior loop.
This rule was not considered in the output ofRNALalifoldand thus has a large impact on the hits obtained from that tool.
Therefore, comparing the output of the above tools against that of version 2.3.5 or less will most likely result in differences.
This version also adds a new software sub-package, RNAlocmin. Detailed
information on this tool can be obtained here. Additionally,
we also add a PDF version of the RNA Bioinformatics tutorial as available
on our homepage to the default instalation target.
The list of all major changes in this version can be found below:
-
Programs
- Print G-Quadruplex corrections in verbose mode of
RNAeval - Change behavior of
RNAfold --outfileoption to something more predictable - Unify max_bp_span usage among sliding window prediction algorithms:
RNAplfold,RNALfold, andRNALalifoldnow consider any base pair (i,j) with (j - i + 1) <= max_bp_span - Add SHAPE reactivity data support to
RNALfold - Add commands-file support for
RNALfold,RNAplfold(hard/soft constraints) - Fix regression for
--canonicalBPonlyswitch inRNAfold/RNAcofold/RNAsubopt - Fix Alidot link in
RNAalifoldmanpage - Restore capability to compile stand-alone
findpathutility
- Print G-Quadruplex corrections in verbose mode of
-
Library
- Add hard constraints to sliding-window MFE and PF implementations
- Add soft constraints to sliding-window MFE and PF implementation for single sequences
- Add SWIG interfaces for sliding-window MFE/PF implementations
- Add proper SWIG interface for alignment and structure plotting functions
- Add proper SWIG interface for duplexfold, duplex_subopt, and its comparative variants
- Add SWIG wrapper for
vrna_exp_params_rescale() - Add explicit destructor for SWIG generated
vrna_md_tobjects - Add SWIG perl5 typemap for simple nested STL vectors
- Add dummy field in
vrna_structured_domains_s - Add SWIG interface for
findpathimplementation - Add
prepare()functions forptype-arrays andvrna_(exp_)param_t - Add warnings for ignored commands in function
vrna_commands_apply() - Add callback featured functions for sliding window MFE and PF implementations
- Change default behavior of adding soft constraints to a
vrna_fold_compound_t(store only) - Several fixes with respect to G-Quadruplex prediction in sliding-window MFE recursions (single sequence and comparative implementation)
- Replace comparative sliding-window MFE recursions (All hits are reported to callback and can be filtered in a post-processing step)
- API: Remove
E_mb_loop_stack()and introduce new functionvrna_E_mb_loop_stack()as a replacement - API: change data type of all constraint bit-flags from
chartounsigned char - API: change data type of
a2sarray in comparative structure prediction fromunsigned shorttounsigned int - API: Change function parameter order in
vrna_probs_window()to follow the style of other callback-aware functions in RNAlib - Move sliding-window MFE implementations to new file
mfe_window.c - Fix redefinition of macro
ON_SAME_STRAND()insubopt.c - Fix dangling end issues in sliding-window MFE implementations
- Fix building sliding-window MFE implementation without SVM support
- Fix parsing of STOCKHOLM 1.0 MSA files that contain MSA spanning multiple blocks
- Fix buffer overflow in hairpin loop sequence motif extraction for circular RNAs
- Fix out-of-bounds memory access in
neighbor.c - Restore capability to use non-standard alphabets for structure prediction
- Restore old-API random number functions in SWIG interface
- Allow additional control characters in MAF MSA input that do not end a block
- Make functions in
pair_mat.hstatic inline - Prevent users from adding out-of-range base pair soft constraints
- Inline print functions in
color_output.inc - Introduce state flag in
vrna_sc_s
-
Package
- Bump
libsvmto version 3.22 - Add
RNAlocmin- Calculate local minima from structures via gradient walks - Add RNA Bioinformatics tutorial (PDF version)
- Add note about SSE optimized code in reference manual
- Fix building PDF Reference manual with non-standard executable paths
- Fix wrong pre-processor flags when enabling single-precision PF computations
- Fix unit testing perl5 interface by including builddir/tests in
PERL5LIBpath - General improvements in reference manual
- Remove obsolete scripts
ct2b.plandcolorrna.plfromsrc/Utilsdirectory - Remove old RNAfold tutorial
- Bump
Version 2.3.x
-
Programs
- Fix duplication of output filename prefix in
RNAfold - Add G-Quadruplex prediction to
RNALalifold - Enable users to turn-off base pair probability computations in
RNAcofoldwith -a option
- Fix duplication of output filename prefix in
-
Library
- Add V3.0 API for sliding window partition function (a.k.a.
RNAPLfold) - Add
SWIGwrappers for callback-based sliding window comparative MFE prediction - Add SSE4.1 multiloop decomposition for single sequence MFE prediction (Thanks to W. B. Langdon)
- Split move set in
neighbor.c
- Add V3.0 API for sliding window partition function (a.k.a.
-
Package
- Enable
RNAfoldunit tests to run in paralllel
- Enable
-
Programs
- Fix double-free when using SHAPE reactivity data in
RNAalifold - Fix z-score output in
RNALfold - Enhance auto-id feature in executable programs
- Always sanitize output file names to avoid problems due to strange FASTA headers
- Lift restrictions of FASTA header length in
RNAfold,RNAcofold, andRNAeval - Add test-suite for
RNAfold - Enable
RNALalifoldto read Clustal/Stockholm/FASTA/MAF alignments in batch-mode - Add parameter option to
RNALalifoldfor colored EPS alignment and structure plot output - Add parameter option to
RNALalifoldto write hits into Stockholm file - Add parameter option to
RNAalifoldto write Stockholm 1.0 formatted output - Add parameter option to
RNAalifoldto suppress stderr spam - Add auto-id feature to
RNAplot,RNALfold,RNAsubopt,RNAplfold,RNAheat - Add SHAPE reactivity derived pseudo-energies as separate output in
RNAalifold - Add colored terminal output to
RNA2Dfold,RNALalifold,RNALfold,RNAduplex,RNAheat,RNAinverse,RNAplfold, andRNAsubopt - Add command line parameters to
RNAsuboptto allow for specification of input/output files
- Fix double-free when using SHAPE reactivity data in
-
Library
- Fix G-Quadruplex global probability computation for single sequences
- Fix out-of-bounds access in
strand_numberarray - Implement sane weighting of SHAPE reactivity data in consensus structure prediction when fewer data than sequences are present
- Substitute field name
A0,B0in data structurevrna_dimer_conc_sbyAc_start,Bc_startto avoid clashes withtermios.h(Mac OSX Python wrapper bug) - Add
ViennaRNA/config.hwith pre-processor definitions of configure time choices - Add functions to procude colored EPS structure alignments
- Add function to write Stockholm 1.0 formatted alignments
- Add function to sanitize file names
- Add callback based implementation for sliding-window MFE prediction (single sequences, comparative structure prediction)
- Add fast API 3.0 implementations to generate structural neighbors and perform steepest descent / random walks
-
Package
- Minimize usage of 'unsafe'
sprintf()calls
- Minimize usage of 'unsafe'
-
Library
- Fix bug in multiloop contributions for comparative partition function
-
Package
- Fix building python2 extension module for OSX
-
Programs
- Fix behavior of
RNAplfoldfor unusually short RNAs - Report SCI of 0 in
RNAalifoldwhen sum of single sequence MFEs is 0
- Fix behavior of
-
Library
- Fix pair probability plist creation with G-Quadruplexes
- Fix init of
vrna_md_tdata structure after call toset_model_details() - Fix bug in consensus partition function with hard constraints that force nucleotides to be paired
- Enable generic hard constraints by default
- Fix init of partition function DP matrices for unusually short RNAs
- Prevent multiple includes of
pair_mat.h
-
Package
- Allow for specification of python2/3-config at configure time
- Fix compilation of functions that use ellipsis/
va_list - Add configure flag to build entirely static executables
-
Programs
- Add unstructured domain feature example to
RNAfoldmanpage
- Add unstructured domain feature example to
-
Library
- Fix regression in
vrna_md_update()that resulted in incomplete init of reverse-basepair type array (mainly affectingRNAalifold) - Fix scaling of secondary structure in EPS plot such that it always fits into bounding box
- Several fixes and improvements for SWIG generated scripting language interface(s)
- Extend coverage of generic hard constraints for partition function computations
- Add missing newline in non-TTY-color output of
vrna_message_info() - Add description for how to use unstructured domains through command files to reference manual
- Fix regression in
-
Package
- Fix compilation issue for Windows platforms with MingW
This version introduces the first transitions towards a more flexible, callback-function based RNA secondary structure decomposition.
The so-called unstructured domains feature
extends the RNA folding grammar with alternative handling
of self-enclosed unpaired stretches in an RNA structure.
This enables one, for instance, to model protein
binding to single stranded sequence motifs, while addressing
the competition between base pairing and binding affinity of
the ligand. The entire extension is not only available using
our RNAlib C-library, but also through the Perl and Python
scripting interfaces. The program RNAfold already includes
a convenience method to add unstructured domain sequence
motifs and their corresponding binding free energy through
easy-to-use command files.
For further details, we refer to our publication "RNA folding with hard and soft constraints", the section on Unstructured Domains and Domain extensions commands of the RNAlib reference manual, and the manpage of RNAfold.
-
Programs
- Introduced command files that subsume constraint definition files (
RNAfoldandRNAcofold) - Replace explicit calls to (non-portable)
asprintf()with portable equivalent functions in the library - Fix regression that prevented several programs to fail on too long input sequences
- Extend EPS dot-plot in
RNAfoldto include motif/binding probabilities from unstructured domains - Add ID manipulation feature to
RNAeval
- Introduced command files that subsume constraint definition files (
-
Library
- Introduce grammar extension through structured and unstructured domains
- Add unstructured domains feature in MFE, partition function, and equilibrium probability computations for single RNA sequences
- Add default implementation for unstructured domains to allow for ligand/protein binding to unpaired structure segments (MFE and PF for single sequences)
- Added utility functions that deal with conversion between different units
- Bugfix in
SWIGwrapped generic soft constraint feature - Add
subopt()andsubopt_zuker()methods to SWIG wrappedfold_compoundobjects - Fix multiloop decomposition in MFE for circular RNAs
- Add separate function to compute pscore for alignments
- Renamed
VRNA_VC_TYPE_*macros toVRNA_FC_TYPE_* - Add variadic functions for error/warning/info message
- Extend API for soft constraint feature for more fine-grained control
- Fix bug in interior loop computations when hard constraints result in non-canonical base pairs
- Restructured reference manual
- Add section on SWIG wrapped functions in reference manual
-
Package
- Fix configure script to deal with situations where
Perlmodule can't be build - Fix bug in
doc/Makefile.amthat prevented HTML installation due to long argument list
- Fix configure script to deal with situations where
Version 2.2.x
-
Programs
- Fix bad memory access that occasionally occured in
RNAsuboptwith sorted output
- Fix bad memory access that occasionally occured in
-
Library
- Fix bad memory access in
vrna_subopt()with sorted output - Do not 'forget' subopt results when output is not written to file handle and sorting is switched off
- Add SWIG wrappers for
vrna_subopt_cb()
- Fix bad memory access in
-
Package
- Correctly show if C11 features are activated in configure status
- Fix autoconf checks to allow for cross compilation again
-
Programs
- Add various new commandline options to manipulate sequence/alignment IDs in
RNAfold,RNAcofoldandRNAalifold - Fix bug for temperature setting in
RNAplfold - Enable
RNAalifoldto use Clustal, Stockholm, FASTA, or MAF alignments as input - Lift restriction of sequence number in alignments for
RNAalifold - Enable ANSI colors for TTY output in
RNAfold,RNAcofold,RNAalifold,RNAsubopt
- Add various new commandline options to manipulate sequence/alignment IDs in
-
Library
- Fix bug in hard constraints usage of exterior loop MFE prediction with odd dangles
- Fix order of hard constraints when read from input file
- Fix several hard constraint corner-cases in MFE and partition function computation when nucleotides must not be unpaired
- Fix bug in partition function scaling for backward compatibility of
ali_pf_fold() - Fix interpretation of
'P'hard constraint for single nucleotides in constraint definition files - Add
'A'command for hard constraints in hard constraints definition files - Stabilize v3.0 API when building RNAlib and third party program linking against it with compilers that use different C/C++ standards
- Use
-fflat-lto-objectsfor static RNAlib library to allow linking without LTO - Add parsers for Clustal, Stockholm, FASTA, and MAF formatted alignment files
- Allow for non-canonical base pairs in MFE and partition function computations if hard constraints demand it
- Enable ANSI colors for TTY output for warnings/errors issued by RNAlib
-
Package
- Add details on how to link against RNAlib to the reference manual
-
Programs
- Fix bad memory access in
RNAalifold - Fix regression in
RNAalifoldto restore covariance contribution ratio determination for circular RNA alignments - Changed output of
RNAsuboptin energy-band enumeration mode to print MFE and energy range in kcal/mol instead of 10cal/mol - Include latest
Kinfoldsources that make use of v3.0 API, therefore speeding up runtime substantially - Re-activate warnings in
RNAevalwhen non-canonical base pairs are encountered
- Fix bad memory access in
-
Library
- Fix syntactic incompatibilities that potentially prevented compilation with compilers other than gcc
- Add function to compare nucleotides encoded in IUPAC format
- Fix regression in energy evaluation for circular RNA sequences
- Fix regression in suboptimal structure enumeration for circular RNAs
- Allow for
P i-j k-lcommands in constraint definition files - Make free energy evaluation functions polymorphic
- Add free energy evaluation functions that allow for specifying verbosity level
- Secure functions in
alphabet.cagainst NULL pointer arguments - Fix incompatibility with
swig≥ 3.0.9 - Fix memory leak in swig-generated scripting language interface(s) for user-provided target language soft-constraint callbacks
- Expose additional functions to swig-generated scripting language interface(s)
- Build Python3 interface by default
- Start of more comprehensive scripting language interface documentation
- Fix linking of python2/python3 interfaces when libpython is in non-standard directory
-
Package
- Restructured
viennarna.specfor RPM based distributions - Several syntactic changes in the implementation to minimize compiler warnings
- Fix
--with-*/--without-*and--enable-*/--disable-*configure script behavior
- Restructured
-
Programs
- Fix partition function scaling for long sequences in
RNAfold,RNAalifold, andRNAup - Fix backtracking issue in
RNAcofoldwhen--noLPoption is activated
- Fix partition function scaling for long sequences in
-
Library
- Fix hard constraints issue for circular RNAs in generating suboptimal structures
-
Package
- Rebuild reference manual only when actually required
-
Programs
- Plugged memory leak in
RNAcofold - Fixed partition function rescaling bug in
RNAup - Fixed bug in
RNALfoldwith window sizes larger than sequence length - Re-added SCI parameter for
RNAalifold - Fixed backtracking issue for large G-quadruplexes in
RNAalifold - Fixed missing FASTA id in
RNAevaloutput - Added option to
RNAalifoldthat allows to specify prefix for output files
- Plugged memory leak in
-
Library
- Several fixes and additional functions/methods in scripting language interface(s)
- Added version information for scripting language interface(s)
- Some changes to allow for compilation with newer compilers, such as
gcc 6.1
-
Programs
-
Fixed regression in
RNAcofoldthat prohibited output of concentration computations -
Fixed behavior of
RNAfoldandRNAcofoldwhen hard constraints create empty solution set (programs now abort with error message)
-
Fixed regression in
-
Library
- Added optional Python 3 interface
-
Added
RNA::ParamsPerl 5 sub-package -
Update
RNA::DesignPerl 5 sub-package - Simplified usage of v3.0 API with default options
- Wrap more functions of v3.0 API in SWIG generated scripting language interfaces
- Plugged some memory leaks in SWIG generated scripting language interfaces
-
Changed parameters of recursion status callback in
vrna_fold_compound_t - Enable definition and binding of callback functions from within SWIG target language
-
Package
-
Added optional subpackage
Kinwalker -
Added several
configureoptions to ease building and packaging under MacOS X -
Added new utility script
RNAdesign.pl
-
Added optional subpackage
-
Programs
-
Fixed bug in
RNAsuboptthat occasionally produced cofolded structures twice
-
Fixed bug in
-
Library
- Removed debugging output in preparations of consensus structure prediction datastructures
-
Programs
-
Added postscipt annotations for found ligand motifs in
RNAfold -
Added more documentation for the constraints features in
RNAfoldandRNAalifold
-
Added postscipt annotations for found ligand motifs in
-
Library
- Restore backward compatibility of get_alipf_arrays()
-
Programs
-
Fix regression bug that occasionally prevented backtracking with
RNAcofold --noLP
-
Fix regression bug that occasionally prevented backtracking with
-
Programs
-
Fix regression bug that made
RNAcofold -aunusable -
Fix regression bug that prohibited
RNAfoldto compute the MEA structure when G-Quadruplex support was switched on -
Fix bug in
Kinfoldto enable loading energy parameters from file -
Fix potential use of uninitialized value in
RNApdist -
Add manpage for
ct2db
-
Fix regression bug that made
-
Library
- Fix MEA computation when G-Quadruplex support is activated
-
Package
-
Allow for
vendor installation of the perl interface usingINSTALLDIRS=vendorat configure time - Install architecture dependent and independent files of the perl and python interface to their correct file system locations
-
Allow for
This version introduces tons of new features. First of all, the new hard- and soft-constraints
allow for easy manipulation of the prediction recursions. For instance, SHAPE reactivity data
can be directly converted to soft-constraints and readily used in MFE, and partition function
computations. Ligand binding to hairpins and interior loops with specific sequence/structure
motifs, e.g. accessible through RNAfold, is another feature that became possible
due to the new constraints framework.
Most of the new features we've implemented into RNAlib require specific new functions that,
partially, replace already existing ones. Therefore, with this version, we start introducing
the new v3.0 API, that will eventually replace the one currently existing.
All functions of the new API are now prefixed with vrna_ that not only makes them
easily recognizable, but also provides some sort of namespacing that helps in the development
of third-party programs linking agains RNAlib.
Through usage of a generic data structure for all secondary structure prediction algorithms, most
functions that implement a special algorithm, such as MFE, or partition function, are now polymorphic.
Thus, there is only one single function in the interface that can deal with multiple variations of
input, such as single sequences or sequence alignments.
However, we will maintain complete backward compatibility until v3 of the ViennaRNA Package.
We also started to make the new API accessible through the Perl and Python
interface, where we introduce a new object-oriented way to use the RNAlib.
Please see the RNAlib Reference Manual, and the individual manpages of the executable programs in the ViennaRNA Package for further details on new features, and feature changes.
-
Programs
- RNAforester is now of version 2.0
- New program RNApvmin to compute pseudo-energy pertubation vector that minimizes discrepancy between observed and predicted pairing probabilities
- SHAPE reactivity support for RNAfold, RNAsubopt, and RNAalifold
- Ligand binding to hairpin- and interior-loop motif support in RNAfold
- New commandline option to limit maximum base pair span for RNAfold, RNAsubopt, RNAcofold, and RNAalifold
- Bugfix in RNAheat to remove numerical instabilities
- Bugfix in RNAplex to allow for computation of interactions without length limitation
- Bugfix in RNAplot for simple layouts and hairpins of size 0
-
Library
- (generic) hard- and soft-constraints for MFE, partition function, base pair probabilities, stochastic backtracking, and suboptimal secondary structures of single sequences, sequence alignments, and sequence dimers
- libsvm version as required for z-scoring in RNALfold is now 3.20
- Stochastic backtracking for single sequences is faster due to usage of Boustrophedon scheme
-
First polymorphic functions
vrna_mfe(),vrna_pf(), andvrna_pbacktrack(). -
The
FLT_OR_DBLmacro is now a typedef - New functions to convert between different secondary structure representations, such as helix lists, and RNAshapes abstractions
- First object-oriented interface for new API functions in the scripting language interfaces
- new ViennaRNA-perl submodule that augments the Perl interface to RNAlib
- Ligand binding to hairpin- and interior-loop motif support in C-library and scripting language interfaces.
-
Package
-
Libraries are generated using
libtool - Linking of libraries and executables defaults to use Link Time Optimization (LTO)
- Large changes in directory structure of the source code files
-
Libraries are generated using
Version 2.1.x
This version fixes a major bug in the energy evaluation of exterior loops and multibranch loops!
In previous versions of the ViennaRNA Package starting with version 2.0, the energy contributions for dangling ends and terminal mismatches of exterior- and multibranch loops were not applied correctly. This problem appeared due to a defective parser that converts the Turner 2004 energy parameters into ViennaRNA code. Therefore, if you compare predictions, e.g. from RNAfold 2.1.9, with those of versions prior to 2.1.9, they may differ. In general, the differences in predicted structures and energies should be very small. However, it may happen that results are totally different.
To assess the impact of the fixed energy parameter usage we re-ran all benchmarks as applied for the
original publication of The ViennaRNA Package 2.0. The resulting performance measures suggest, that the
impact is actually very small, although predictions seem to be slightly better with version 2.1.9.
See the ViennaRNA Package 2.0 - Performance page for details.
-
Programs
- Fixed integer underflow bug in RNALfold
- Added Sequence Conservation index (SCI) option to RNAalifold
-
Library
- Fixed bug in energy evaluation of dangling ends / terminal mismatches of exterior loops and multibranch loops
- Fixed bug in alifold partition function for circular RNAs
- Fixed bug in alifold that scrambled backtracing with activated G-Quadruplex support
- Fixed bug in alifold backtracking for larger G-Quadruplexes
-
Utils
- Avoid warning for missing sequence constraint in switch.pl
-
Programs
- Repaired incorporation of RNAinverse user provided alphabet
- Fix missing FASTA ID in RNAeval output
-
Library
- prevent race condition in parallel calls of Lfold()
- Fixed memory bug in Lfold() that occured using long sequences and activated G-Quad support
-
Utils
- Added latest version of switch.pl
-
Programs
- Fixed bug in RNALfold -z
-
Library
- Python and Perl interface are compiling again under MacOSX. See Install Notes.
- Fixed handling of C arrays in Python interface
-
Utils
- Added latest version of switch.pl
- Make relplot.pl work with RNAcofold output
-
Programs
- New commandline switches allow for elimination of non-canonical base pairs from constraint structures in RNAfold, RNAalifold and RNAsubopt
-
Library
- updated moveset functions
- final fix for discrepancy of tri-loop evaluation between partition function and mfe
- pkg-config file now includes the OpenMP linker flag if necessary
-
Utils
- New program ct2db allows for conversion of .ct files into dot-bracket notation (incl. pseudo-knot removal)
-
Library
- Fix for discrepancy between special hairpin loop evaluation in partition functions and MFE
-
Library
- Fix of G-quadruplex support in subopt()
- Fix for discrepancy between special hairpin loop evaluation in partition functions and MFE
-
Programs
- RNAfold: Bugfix for ignoring user specified energy parameter files
- RNAcofold: Bugfix for crashing upon constrained folding without specifying a constraint structure
- RNAsubopt: Added G-quadruplex support
- RNAalifold: Added parameter option to specify base pair probability threshold in dotplot
-
Library
- Fix of several G-quadruplex related bugs
- Added G-quadruplex support in subopt()
-
Programs
- RNAfold: Bugfix for randomly missing probabilities in dot-plot during batch job execution
- RNAeval: Bugfix for misinterpreted G-quadruplex containing sequences where the quadruplex starts at nucleotide 1
- RNAsubopt: Slight changes to the output of stochastic backtracking and zuker subopt
-
Library
- Fix of some memory leaks
- Bugfixes in zukersubopt(), assign_plist_from_pr()
- New threadsafe variants of putoutpU_prob*() for LPfold()
- Provision of python2 interface support. This interface is very basic at the moment and the configure script default is to disable it. If you want to try it out, use it at your own risk. Comments and remarks about usability are welcome.
-
Library
- Bugfix to restore backward compatibility with ViennaRNA Package 1.8.x API (this bug also affected proper usage of the the perl interface)
The ViennaRNA Package was recently extended to take a certain class of higher-order structure motifs, so called G-quadruplexes, into account, too.
This is the recursion scheme that we use to predict these structure motifs:
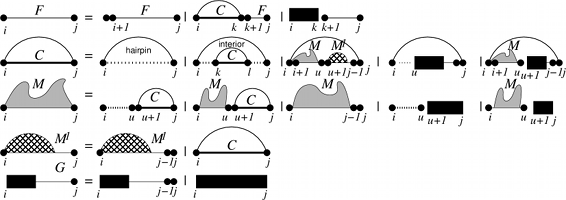
G-quadruplex predictions are marked by the character "+" in the extended dot-parenthesis notation output. Thus, RNAfold output may look like
$ echo "GGCUGGUGAUUGGAAGGGAGGGAGGUGGCCAGCC" | Progs/RNAfold -g
GGCUGGUGAUUGGAAGGGAGGGAGGUGGCCAGCC
((((((..........++.++..++.++)))))) (-21.39)
Calculation of the base pair probabilities for the above example then results in a dot-plot like this
-
Programs
- G-Quadruplex support in RNAfold, RNAcofold, RNALfold, RNAalifold, RNAeval and RNAplot
This introduces a new commandline option '--gquad/-g' that enables G-quadruplex prediction. The dot-bracket notation for the structure output now contains an additional character '+' that indicates the presence of such a G-tetrad. Consecutive '+' signs always come in sets of 4 interspaced by some '.', marking the four G-runs and its linker sequences that comprise the quadruplex, respectively. - changes in RNAPKplex behavior and commandline options
- new features and bugfixes in RNAplex and RNAsnoop
- warning messages for all programs if an unsupported dangle option is selected
- G-Quadruplex support in RNAfold, RNAcofold, RNALfold, RNAalifold, RNAeval and RNAplot
-
Library
- LPfold got a new option to output its computations in split-mode
- several G-Quadruplex related functions were introduced with this release
- several functions for moves in an RNA landscape were introduced
- new function in alipfold.c now enables access to the partition function matrices of alipf_fold()
- different numeric approach was implement for concentration dependend co-folding to avoid instabilities which occured under certain circumstances
-
Utils
- To enable fast plotting of RNA2Dfold output we now also provide two gri-scripts
"2Dlandscape_mfe.gri" and "2Dlandscape_pf.gri".
To use them, simply redirect the output of RNA2Dfold into a text file and then call the appropriate script with the RNA2Dfold output filename as first parameter, e.g.:This will create a postscript file named "my_2Dfold_output.txt.ps" that displays the MFE output in a nice landscape like manner$ ./2Dlandscape_mfe.gri my_2Dfold_output.txt
- The Utils script "relplot.pl" got a new commandline option "-a" that enables annotation of the accessibility in the structure plot
- To enable fast plotting of RNA2Dfold output we now also provide two gri-scripts
"2Dlandscape_mfe.gri" and "2Dlandscape_pf.gri".
-
Package
- The auxilary energy parameter files in the source tree now reside in the 'misc' directory.
They are, however, still installed at the same locations as in previous versions of the Package. - Some changes in the build process now enable easier cross compilation of the Package
- The example sourcecode in the reference manual got an update to comply with the specifications of ViennaRNA Package 2.x
- The auxilary energy parameter files in the source tree now reside in the 'misc' directory.
Version 2.0.x
-
Programs
- Bugfix for RNAplfold where segfault happened upon usage of -O option
- Corrected misbehavior of RNAeval and RNAplot in tty mode
-
Programs
- Bugfix for bad type casting with gcc under MacOSX (resulted in accidental "sequence too long" errors)
- Bugfix for disappearing tri-/hexaloop contributions when read in from certain parameter files
- Bugfix for RNALfold that segfaulted on short strange sequences like AT+ repeats
- Change of RNA2Dfold output format for stochastic backtracking
-
Programs
- Restored z-score computation capabilities in RNALfold
-
Programs
- Bugfix for RNAcofold partition function
-
Library
- Perl wrapper compatibility to changed RNAlib has been restored
- Backward compatibility for partition function calls has been restored [under certain circumstances under-/overflows in the calculations of pf_fold(), alipf_fold(), co_pf_cold() and pfl_fold() appeared in version 2.0.3]
-
Programs
- Bugfix for RNAalifold partition function and base pair probabilities in v2.0.3b
- Added Boltzmann factor scaling in RNAsubopt, RNAalifold, RNAplfold and RNAcofold
-
Library
- Bugfix for alipfold() in v2.0.3b
- Restored threadsafety of folding matrix access in LPfold.c, alipfold.c, part_func.c, part_func_co.c and part_func_up.c
- Added several new functions regarding threadsafe function calls in terms of concurrently
changing the model details. The new functions with the suffix '_par' may now be used to
pass an energy contribution datastructure filled with additional information about the model
details. In this way, seperation from the global variables
dangles, noLonelyPairs, noGU, no_closingGU, tetra_loop, temperature, logML
is possible. Furthermore, the new functions also provide detachement from the global variablesdo_backtrack, fold_constrained
This works for all implementations in
- fold.c
- part_func.c
- LPfold.c
- alipfold.c
Basic support is provided in
- cofold.c
- part_func_co.c
- subopt.c
However, since all cofolding functions currently rely on the global variable 'cut_point', they are not considered really threadsafe yet, i.e. a change in the cut_point while concurrent calculations take place will result in unpredictable results!
Threadsafety of the remaining functions, regarding the above mentioned global variables will be incoorporated in upcoming versions
See the reference manual of the RNAlib for more details (params.h, part_func.h, etc.)
-
Package
- Added pkg-config file in the distribution to allow easy checks for certain RNAlib2
versions, compiler flags and linker flags. This makes linking of third party programs
agains RNAlib2 much more easier.
E.g. a call of 'pkg-config --modversion RNAlib2' reveals the version of the installed RNAlib. Developers who use autoconf may now take advantage of the different PKG_ macros to retrieve version, linker and compiler flags
- Added pkg-config file in the distribution to allow easy checks for certain RNAlib2
versions, compiler flags and linker flags. This makes linking of third party programs
agains RNAlib2 much more easier.
-
Programs
- added support for Boltzmann factor scaling in RNAfold
- fixed fastaheader to filename bug
-
Library
- plugged some memory leaks
-
Package
- First official release of version 2.0
- included latest bugfixes
History
-
09.05.2011
Fixed file name issue that occured under HFS on MacOS X (unbelievably HFS is not capable to distinguish files solely based on the case of the letters in the filename)
-
18.04.2011
Fixed RNAcofold bug (memory corruption when two complementary sequences form a full length helix)
-
17.03.2011
Fixed RNAsubopt bug (invalid free() appeared sometimes depending on compiler used)
-
10.03.2011
Replaced calls to non-ANSI function strndup() to ensure compatibility with MacOS X and FreeBSD
-
07.03.2011
Fixed RNAup bug (interaction mode might have taken wrong sequence as target in batch mode)



