 |
 |
 |
 |
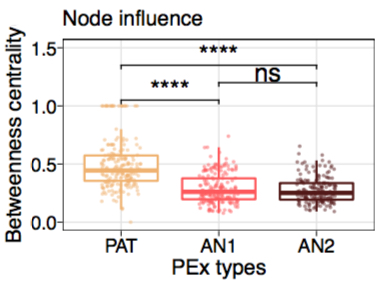 Microbial community organization designates distinct pulmonary exacerbation types and predicts treatment outcome in cystic fibrosis. S Widder, K Opron, L Carmody, L Kalikin, L Caverly, J LiPuma. Nature Communications 2024 Microbial community organization designates distinct pulmonary exacerbation types and predicts treatment outcome in cystic fibrosis. S Widder, K Opron, L Carmody, L Kalikin, L Caverly, J LiPuma. Nature Communications 2024
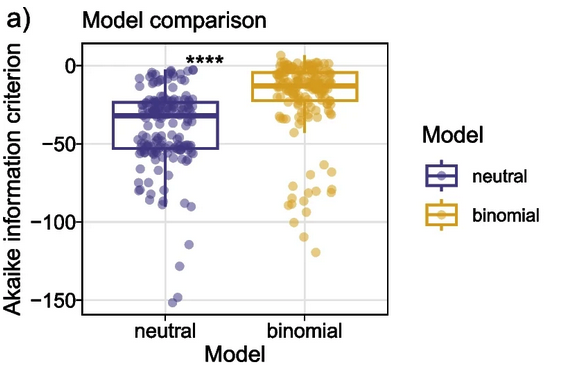 Longitudinal Microbial and Molecular Dynamics in the Cystic Fibrosis Lung after Elexacaftor-Tezacaftor-Ivacaftor therapy. C Martin, D Guzior, C Gonzalez, M Okros, et al. Respiratory Research 2023 Longitudinal Microbial and Molecular Dynamics in the Cystic Fibrosis Lung after Elexacaftor-Tezacaftor-Ivacaftor therapy. C Martin, D Guzior, C Gonzalez, M Okros, et al. Respiratory Research 2023
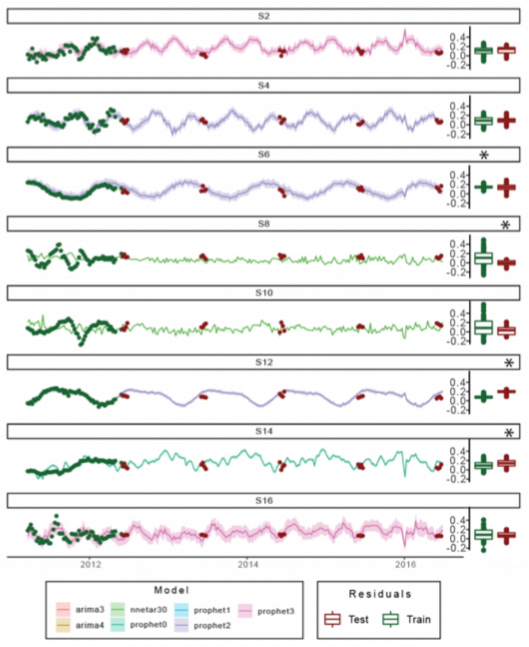 Forecasting the dynamics of a complex microbial community using integrated meta-omics. F Delogu, B Kunath, P Queiros, R Halder, L Lebrun, P Pope, P May, S Widder, E Muller, P Wilmes. Nature Ecology Evolution 2023 Forecasting the dynamics of a complex microbial community using integrated meta-omics. F Delogu, B Kunath, P Queiros, R Halder, L Lebrun, P Pope, P May, S Widder, E Muller, P Wilmes. Nature Ecology Evolution 2023
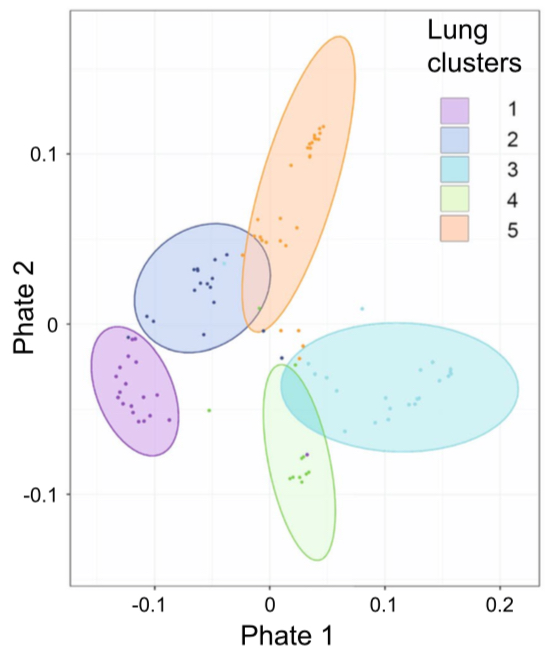 Metagenomic sequencing reveals time, host, and body compartment-specific viral dynamics after lung transplantation. S Widder, I Goerzer, B Friedel et al. and E Puchammer-Stoeckl. Microbiome 2022 Metagenomic sequencing reveals time, host, and body compartment-specific viral dynamics after lung transplantation. S Widder, I Goerzer, B Friedel et al. and E Puchammer-Stoeckl. Microbiome 2022
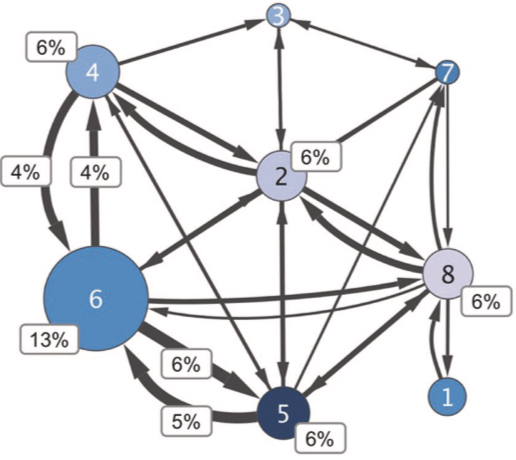 Association of bacterial community types, functional microbial processes and lung disease in cystic fibrosis airways. S Widder, J Zhao, L Carmody, et al. and J J LiPuma. ISMEJ 2021 Association of bacterial community types, functional microbial processes and lung disease in cystic fibrosis airways. S Widder, J Zhao, L Carmody, et al. and J J LiPuma. ISMEJ 2021
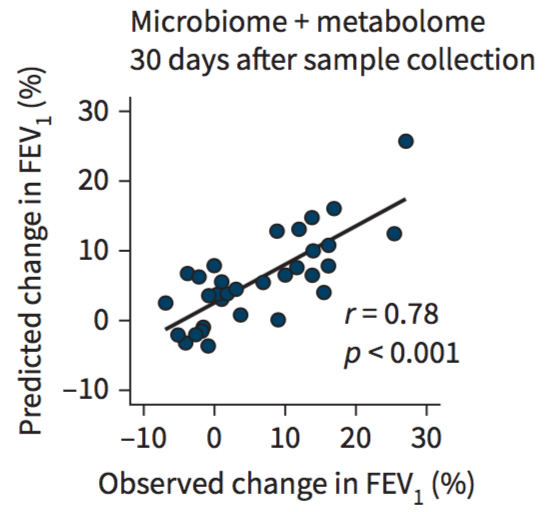 Multi-omics profiling predicts allograft function after lung transplantations. M Watzenboeck, A Gorki, F Quattrone, R Gawish et al. and S Widder & S Knapp. European Respiratory Journal 2021 Multi-omics profiling predicts allograft function after lung transplantations. M Watzenboeck, A Gorki, F Quattrone, R Gawish et al. and S Widder & S Knapp. European Respiratory Journal 2021
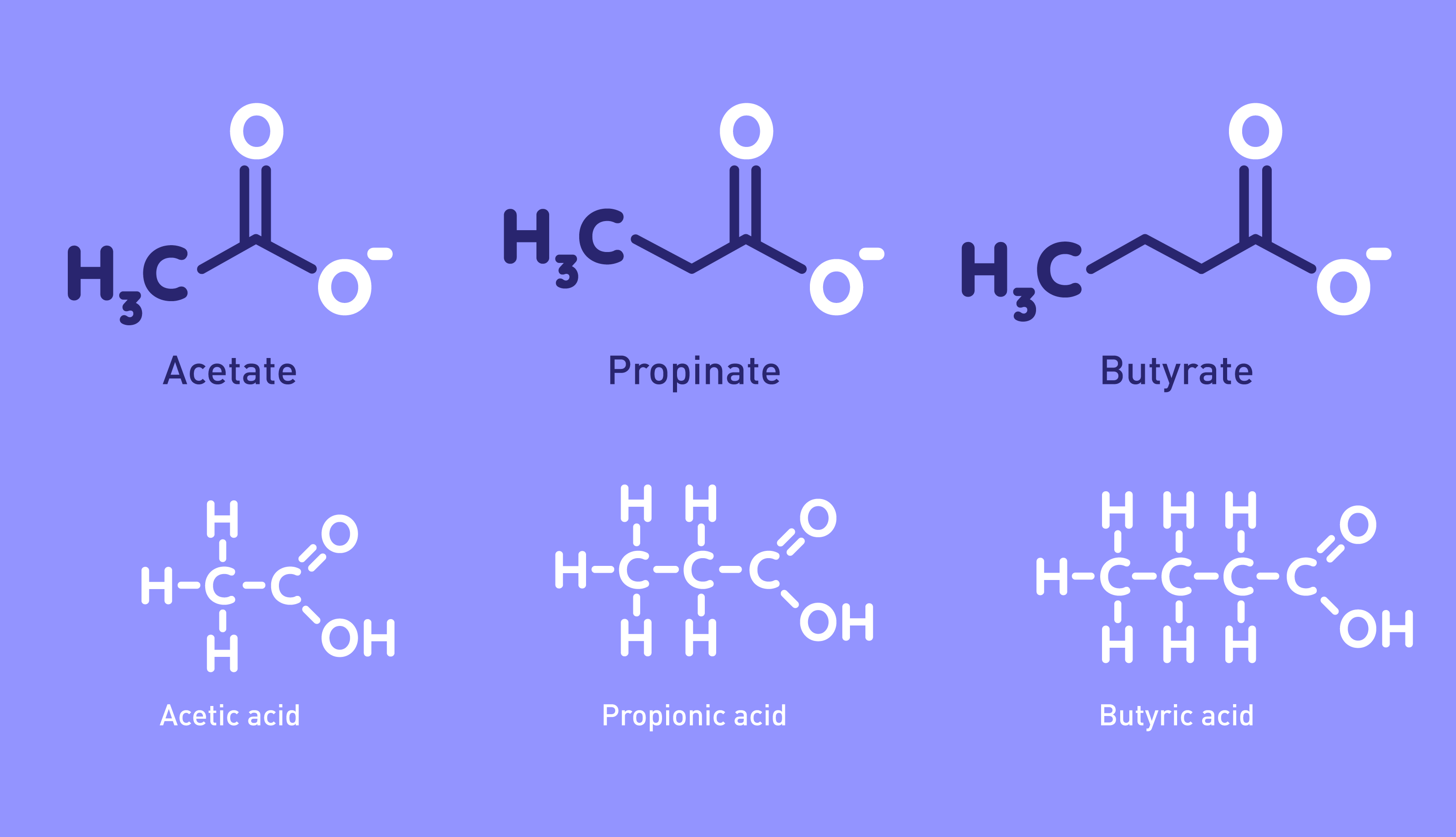 Editorial: Microbial Metabolites in Cystic Fibrosis: A Target for Future Therapy? S Widder and S Knapp. American Journal of Respiratory Cell and Molecular Biology 2019 Editorial: Microbial Metabolites in Cystic Fibrosis: A Target for Future Therapy? S Widder and S Knapp. American Journal of Respiratory Cell and Molecular Biology 2019
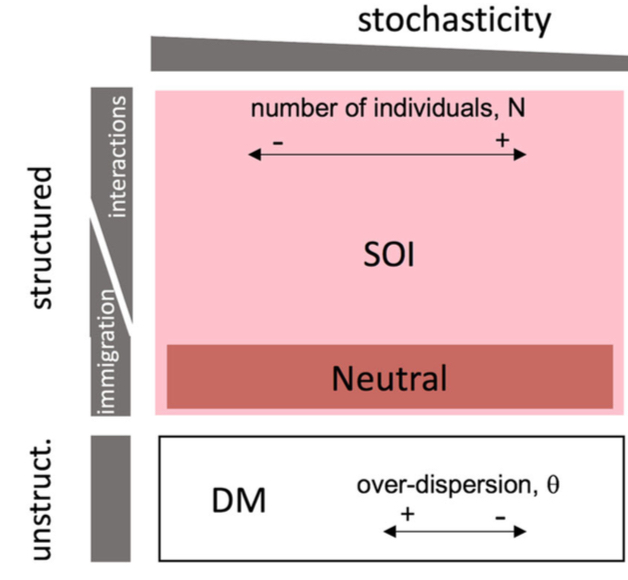 Signatures of ecological processes in microbial community time series. K Faust, F Bauchinger, B Laroche, S de Buyl, L Lahti, A Washburne, D Gonze and S Widder. Microbiome 2018 Signatures of ecological processes in microbial community time series. K Faust, F Bauchinger, B Laroche, S de Buyl, L Lahti, A Washburne, D Gonze and S Widder. Microbiome 2018
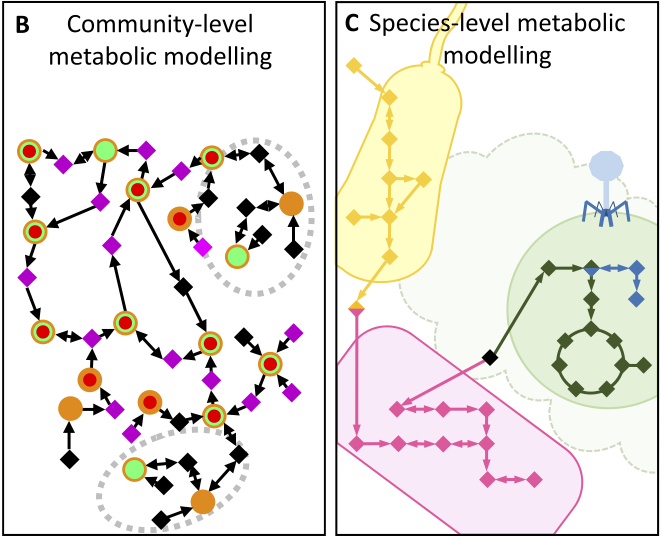 Using metabolic networks to resolve ecological properties of microbiomes. E Muller, K Faust, S Widder, M Herold, S Martinez Abas and P Wilmes. Cur Opin Sys Biol. 2017 Using metabolic networks to resolve ecological properties of microbiomes. E Muller, K Faust, S Widder, M Herold, S Martinez Abas and P Wilmes. Cur Opin Sys Biol. 2017
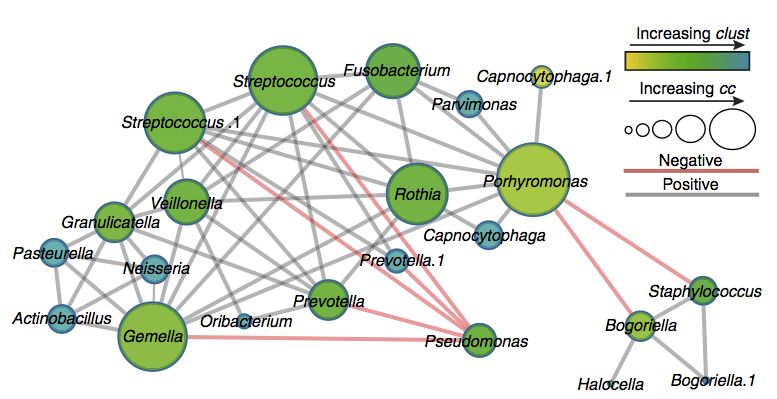 Ecological networking of cystic fibrosis lung infections. Quinn RA, Whiteson K, Lim YW, Zhao J, Conrad D, LiPuma JJ, Rohwer F, Widder S. NPJ Biofilms Microbiomes. 2016 Dec Ecological networking of cystic fibrosis lung infections. Quinn RA, Whiteson K, Lim YW, Zhao J, Conrad D, LiPuma JJ, Rohwer F, Widder S. NPJ Biofilms Microbiomes. 2016 Dec
 The origin and evolution of cell types. Arendt D*, Musser JM*, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, Wagner GP. Nat Rev Genet. 2016 DecNews and Features: Science (350/2015): In depth-Using evolution to better identify cell types The origin and evolution of cell types. Arendt D*, Musser JM*, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, Wagner GP. Nat Rev Genet. 2016 DecNews and Features: Science (350/2015): In depth-Using evolution to better identify cell types
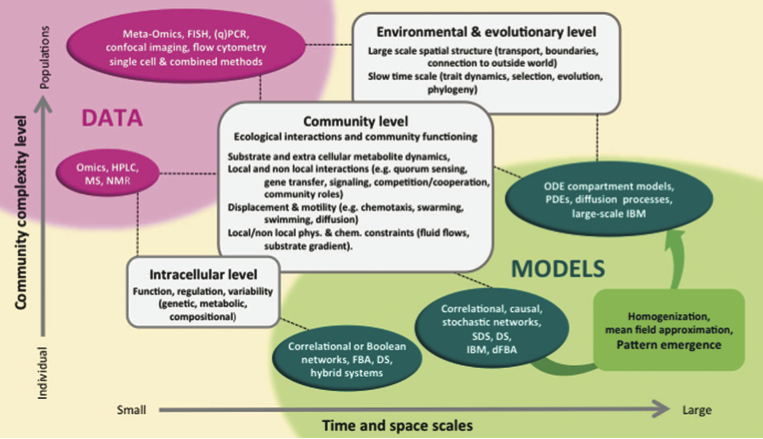 Challenges in microbial ecology: building predictive understanding of community function and dynamics. Widder S, Allen R, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, Cordero OX, Brown SP, Momeni B, Shou W, Kettle H, Flint HJ, Haas AF, Laroche B, Kreft JU, Rainey PB, Freilich S, Schuster S, Milferstedt K, van der Meer JR, Groβkopf T, Huisman J, Free A, Picioreanu C, Quince C, Klapper I, Labarthe S, Smets BF, Wang H, Isaac Newton Institute Fellows, Soyer OS. ISME J. 2016 Nov Challenges in microbial ecology: building predictive understanding of community function and dynamics. Widder S, Allen R, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, Cordero OX, Brown SP, Momeni B, Shou W, Kettle H, Flint HJ, Haas AF, Laroche B, Kreft JU, Rainey PB, Freilich S, Schuster S, Milferstedt K, van der Meer JR, Groβkopf T, Huisman J, Free A, Picioreanu C, Quince C, Klapper I, Labarthe S, Smets BF, Wang H, Isaac Newton Institute Fellows, Soyer OS. ISME J. 2016 Nov
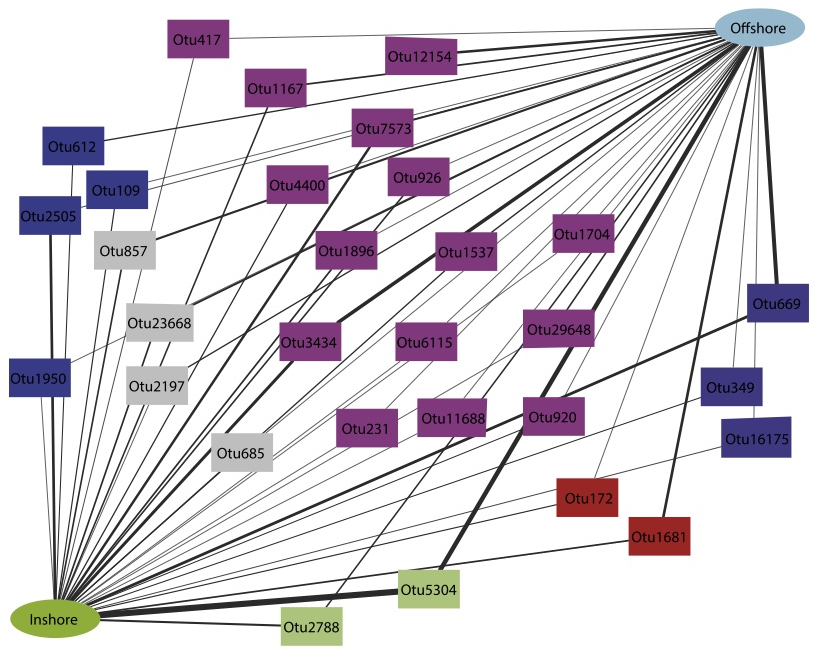 Biogeographic variation in the microbiome of the ecologically important sponge, Carteriospongia foliascens. Luter HM, Widder S, Botté ES, Abdul Wahab M, Whalan S, Moitinho-Silva L, Thomas T, Webster NS. PeerJ. 2015 Dec Biogeographic variation in the microbiome of the ecologically important sponge, Carteriospongia foliascens. Luter HM, Widder S, Botté ES, Abdul Wahab M, Whalan S, Moitinho-Silva L, Thomas T, Webster NS. PeerJ. 2015 Dec
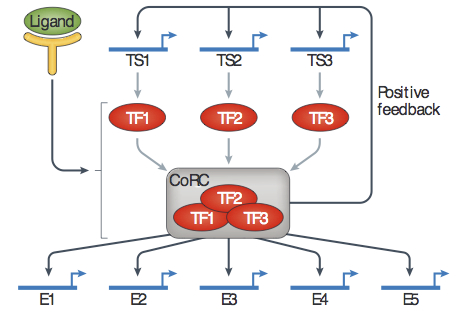
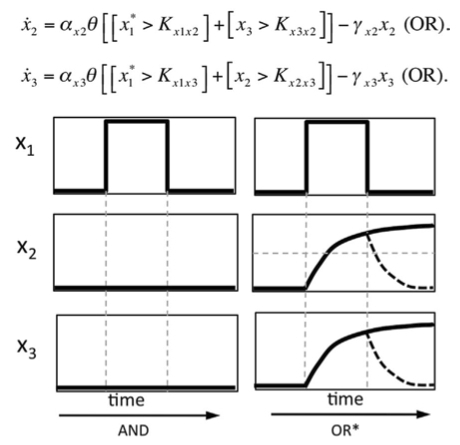 Wiring for Independence: Positive Feedback Motifs Facilitate Individuation of Traits in Development and Evolution. Pavlicev M, Widder S. J Exp Zool B Mol Dev Evol. 2015 Mar Wiring for Independence: Positive Feedback Motifs Facilitate Individuation of Traits in Development and Evolution. Pavlicev M, Widder S. J Exp Zool B Mol Dev Evol. 2015 Mar
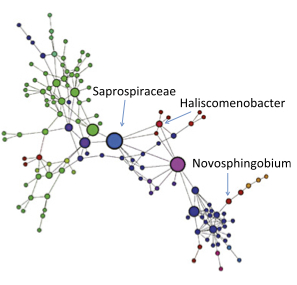 Fluvial network organization imprints on microbial co-occurrence networks. Widder S, Besemer K, Singer GA, Ceola S, Bertuzzo E, Quince C, Sloan WT, Rinaldo A, Battin TJ. Proc Natl Acad Sci U S A. 2014 Sep News and Features:
PNAS (111/2014): This week in PNAS Fluvial network organization imprints on microbial co-occurrence networks. Widder S, Besemer K, Singer GA, Ceola S, Bertuzzo E, Quince C, Sloan WT, Rinaldo A, Battin TJ. Proc Natl Acad Sci U S A. 2014 Sep News and Features:
PNAS (111/2014): This week in PNAS
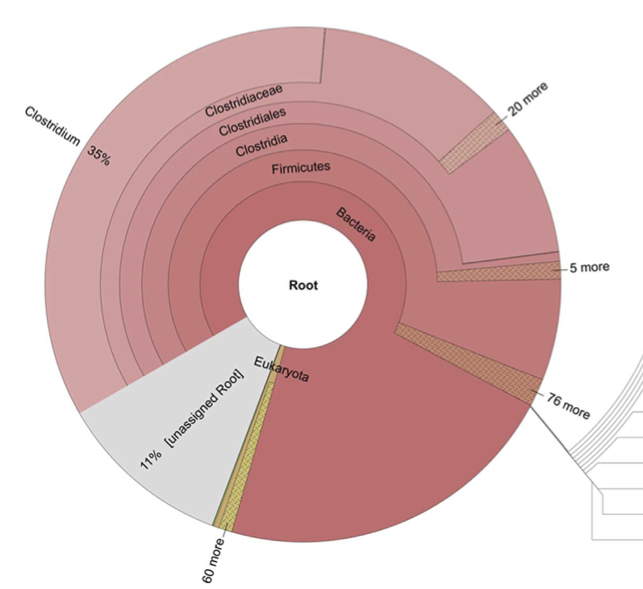 Metagenomic analysis reveals presence of Treponema denticola in a tissue biopsy of the Iceman. Maixner F*, Thomma A*, Cipollini G, Widder S, Rattei T, Zink A. PLoS One. 2014 Jun News and Features:Der Kurier, Printversion (16/7/2014): Schon Ötzi wurde von Zahnfleischentzündung gequält. Der Standard (16/7/2014): Ötzis Knochenanalyse enthüllt "nichtmenschliche" DNA Universität Wien - uni:view magazin (15/7/2014): Ötzis "nichtmenschliche" DNA entschlüsselt Metagenomic analysis reveals presence of Treponema denticola in a tissue biopsy of the Iceman. Maixner F*, Thomma A*, Cipollini G, Widder S, Rattei T, Zink A. PLoS One. 2014 Jun News and Features:Der Kurier, Printversion (16/7/2014): Schon Ötzi wurde von Zahnfleischentzündung gequält. Der Standard (16/7/2014): Ötzis Knochenanalyse enthüllt "nichtmenschliche" DNA Universität Wien - uni:view magazin (15/7/2014): Ötzis "nichtmenschliche" DNA entschlüsselt
 Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Berry D, Widder S. Front Microbiol. 2014 May Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Berry D, Widder S. Front Microbiol. 2014 May
 Ultra deep sequencing of Listeria monocytogenes sRNA transcriptome revealed new antisense RNAs. Behrens S, Widder S, Mannala GK, Qing X, Madhugiri R, Kefer N, Abu Mraheil M, Rattei T, Hain T. PLoS One. 2014 Feb Ultra deep sequencing of Listeria monocytogenes sRNA transcriptome revealed new antisense RNAs. Behrens S, Widder S, Mannala GK, Qing X, Madhugiri R, Kefer N, Abu Mraheil M, Rattei T, Hain T. PLoS One. 2014 Feb
 Evolvability of feed-forward loop architecture biases its abundance in transcription networks. Widder S, Solé R, Macía J. BMC Syst Biol. 2012 Jan Evolvability of feed-forward loop architecture biases its abundance in transcription networks. Widder S, Solé R, Macía J. BMC Syst Biol. 2012 Jan
 Specialized or flexible feed-forward loop motifs: a question of topology. Macía J*, Widder S*, Solé R. BMC Syst Biol. 2009 Aug Specialized or flexible feed-forward loop motifs: a question of topology. Macía J*, Widder S*, Solé R. BMC Syst Biol. 2009 Aug
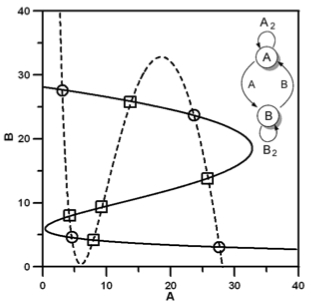 Why are cellular switches Boolean? General conditions for multistable genetic circuits. Macía J, Widder S, Solé R. J Theor Biol. 2009 Nov Why are cellular switches Boolean? General conditions for multistable genetic circuits. Macía J, Widder S, Solé R. J Theor Biol. 2009 Nov
 Monomeric bistability and the role of autoloops in gene regulation. Widder S*, Macía J*, Solé R. PLoS One. 2009 Monomeric bistability and the role of autoloops in gene regulation. Widder S*, Macía J*, Solé R. PLoS One. 2009
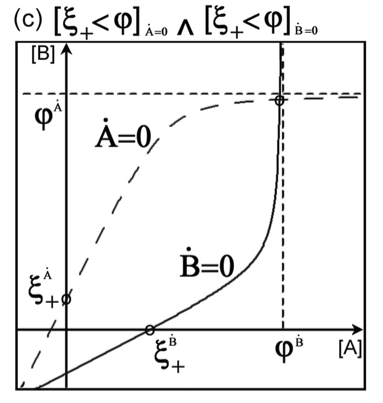
 Dynamic patterns of gene regulation I: simple two-gene systems. Widder S, Schicho J, Schuster P. Front Microbiol. 2014 May Dynamic patterns of gene regulation I: simple two-gene systems. Widder S, Schicho J, Schuster P. Front Microbiol. 2014 May
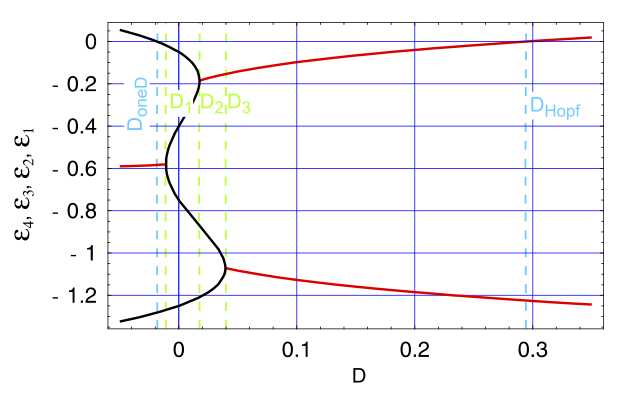
 A minimal and self-consistent in silico cell model based on macromolecular interactions. Flamm C, Endler L, Müller S, Widder S, Schuster P. Philos Trans R Soc Lond B Biol Sci. 2007 Oct A minimal and self-consistent in silico cell model based on macromolecular interactions. Flamm C, Endler L, Müller S, Widder S, Schuster P. Philos Trans R Soc Lond B Biol Sci. 2007 Oct
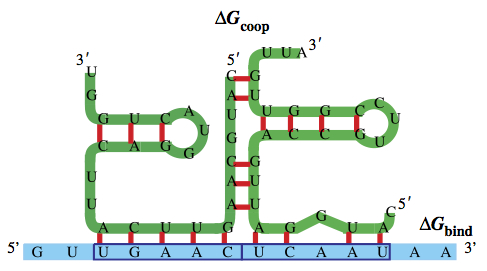
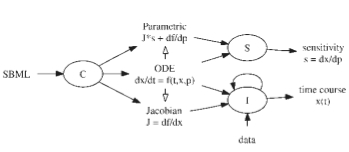 The SBML ODE Solver Library: a native API for symbolic and fast numerical analysis of reaction networks. Machné R, Finney A, Müller S, Lu J, Widder S, Flamm C. Bioinformatics. 2006 Jun The SBML ODE Solver Library: a native API for symbolic and fast numerical analysis of reaction networks. Machné R, Finney A, Müller S, Lu J, Widder S, Flamm C. Bioinformatics. 2006 Jun
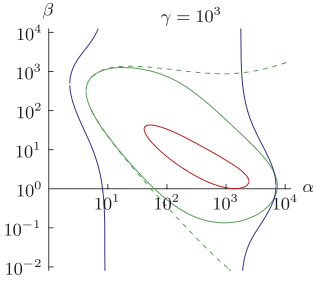 A generalized model of the repressilator. Müller S, Hofbauer J, Endler L, Flamm C, Widder S, Schuster P. Journal Math Biol. 2006 Dec A generalized model of the repressilator. Müller S, Hofbauer J, Endler L, Flamm C, Widder S, Schuster P. Journal Math Biol. 2006 Dec
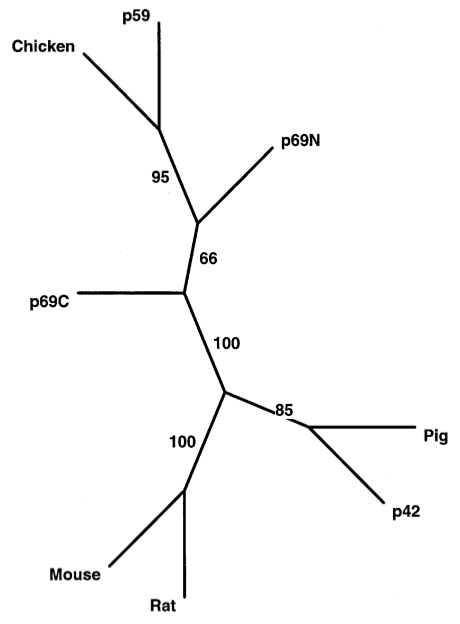 p59OASL, a 24–54 oligoadenylate synthetase like protein: a novel human gene related to the 24–54 oligoadenylate synthetase family. Hartmann R, Olsen HS, Widder S, Jørgensen R, Justesen J. Nucleic Acids Research, 1998 p59OASL, a 24–54 oligoadenylate synthetase like protein: a novel human gene related to the 24–54 oligoadenylate synthetase family. Hartmann R, Olsen HS, Widder S, Jørgensen R, Justesen J. Nucleic Acids Research, 1998
| |