Adaptive search
 RNAsubopt (part of the ViennaRNA package), in its Boltzman sampling nature, usually produces only structures very near to the global minimum over and over again.
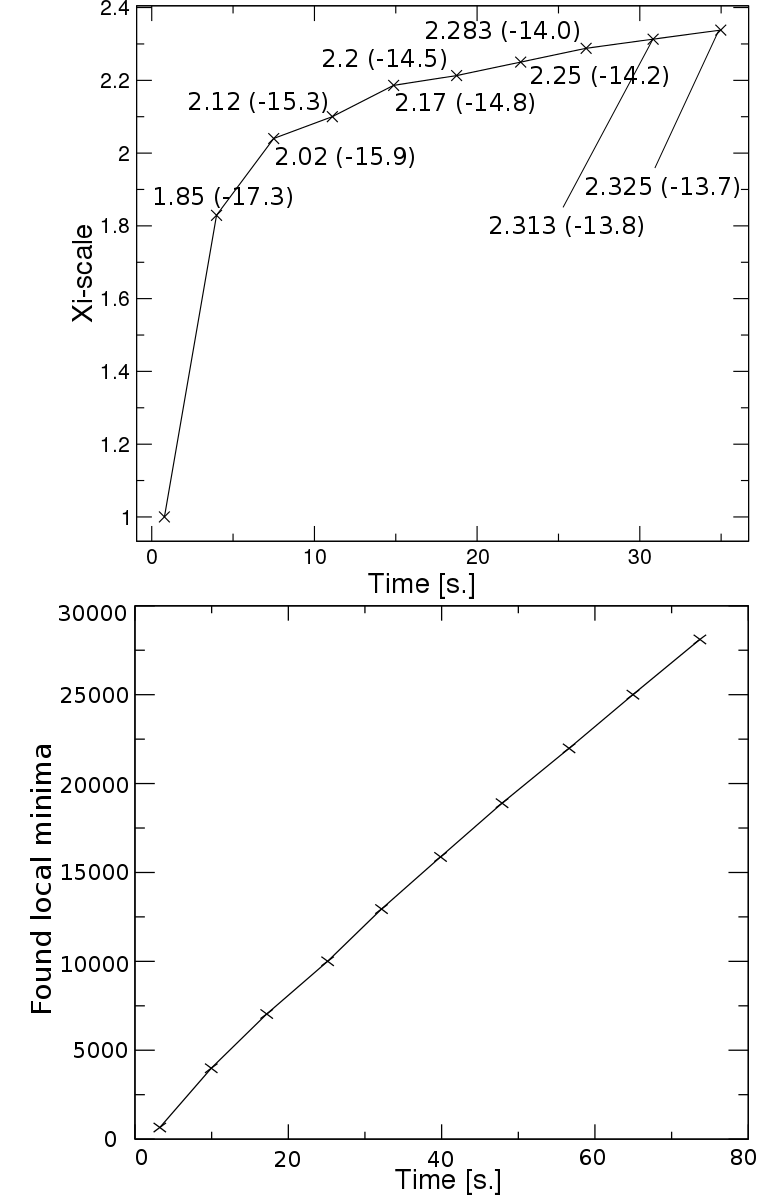
Generating samples with higher energy can be done by introducing a scaling parameter (xi) in the sampling, which can be seen as an alteration of the sampling temperature.
The adaptive search schedule aims to find local minima, that cover a desired energy range from the global optimum. For this purpose, a criterion of coverage has been introduced
to be sure that we have successfully covered local minima up to some energy. After this, sampling shifts to higher energies by increasing the scaling parameter, as long as the desired
energy range is not covered or a maximal number of iterations is not reached.
RNAsubopt (part of the ViennaRNA package), in its Boltzman sampling nature, usually produces only structures very near to the global minimum over and over again.
Generating samples with higher energy can be done by introducing a scaling parameter (xi) in the sampling, which can be seen as an alteration of the sampling temperature.
The adaptive search schedule aims to find local minima, that cover a desired energy range from the global optimum. For this purpose, a criterion of coverage has been introduced
to be sure that we have successfully covered local minima up to some energy. After this, sampling shifts to higher energies by increasing the scaling parameter, as long as the desired
energy range is not covered or a maximal number of iterations is not reached.
The adaptive search schedule script is written in Python 2.7 and requires both programs RNAsubopt and RNAlocmin to work.
To make it run, just download the script asearch_2.py, edit the paths to programs on the top of the script, and run python asearch_2.py -I input_file.
The output is generated in the same directory as the input file with filename extensions .lm (basic output - list of generated local minima), .lmns (list of local minima after filtering), and .bhg (BHGgraph if --bhg option was specified).
Run python asearch_2.py -h to get help and available options.
First, it constructs xi-to-energy table, where the average energy values for different scale parameters are stored. (New version constructs this table already on the run.) Then it iteratively runs RNAsubopt and RNAlocmin and vary the scaling parameter to obtain the best set of local minima. The coverage criterion can be modified with a coverage parameter, which is by default set to 0.1. Redefining it to a higher number shifts the sampling to higher energies much sooner, and vice versa. We recommend using values between 0.05 to 0.8. You can find more information in the article below.
In case you are using our software for your publications you may want to cite:
Basin Hopping Graph: A computational framework to characterize RNA folding landscapes
Marcel Kucharík, Ivo L. Hofacker, Peter F. Stadler and Jing Qin
Bioinformatics 2014
doi:10.1093/bioinformatics/btu156
bibtex

 tbi.univie.ac.at -- programming
tbi.univie.ac.at -- programming